
BRD4 regulates the induction and maintenance of cancer stem cells in squamous cell carcinoma
Non-melanoma skin cancer (NMSC) is one of the most frequently diagnosed cancers across the world (1). Greater than 70% of NMSCs are basal cell carcinomas (BCC) and about 25% are squamous cell carcinomas (SCCs). Both forms have good prognosis if detected and treated early (2). The majority of NMSC-related deaths (~75%) are due to SCC, which has a mortality rate (MR) of up to about 10%; MR in BCC is about 0.02% (1,2). SCC represents an aggressive group of cancers that arise due to uncontrolled growth of squamous epithelial cells in the head and neck region, oral cavity, esophagus, lungs, bladder or cervix. SCC has considerable metastatic potential and high risk of recurrence owing to the presence of highly proliferative and invasive squamous cells (2). Besides surgery, cryosurgery and radiotherapy, different combinations of therapeutic agents including cisplatin, 5-fluorouracil, doxorubicin, epidermal growth factor receptor inhibitors (cetuximab, panitumumab and matuzumab) as well as anti-programmed cell death protein 1 (PD-1) immune checkpoint inhibitors (cemiplimab and pembrolizumab) are used to treat various forms of SCC. However, the therapeutic efficacy of these drugs remains limited by side effects, chemoresistance and, perhaps most importantly, increased risk of recurrence (3). The possibility that drugs in use to treat SCC often miss the very small but all-important cancer stem cell (CSC) populations has been previously entertained as one of the reasons behind chemotherapy-related relapse in SCC (4). Therefore, investigating whether SCCs harbor CSCs and identifying targets of potential therapeutic utility against such small cell populations is of paramount importance.
In this context, a recent study by Fisher et al. (5) revealed that CSCs displaying an aggressive phenotype with significant migration, invasion and growth potential can be isolated from spheroid cultures of SCC cell lines. The enriched CSC population they identified expressed high levels of the bromodomain and extra terminal (BET) domain family member BRD4, the enhancer of zeste homologue 2 (EZH2), the P53 family member P63 splice variant ΔNp63α, the SRY-box transcription factor 2 (SOX2) and the octamer-binding transcription factor 4 (OCT4) among others. The authors used small molecule inhibitors and clustered regularly interspaced short palindromic repeats (CRISPR)-based gene depletion and rescue approaches to investigate the role of BRD4, EZH2, the signal transducer and activator of transcription 3 (STAT3) and P63 in the induction and maintenance of CSCs both in vitro and in nude mice xenografted with genetically-manipulated SCCs. The study showed that BRD4 regulates the transcriptional activity of ΔNp63α in SCC cells with a stem cell (SC)-like phenotype through the recruitment of EZH2/STAT3 complexes, presumably to the promotor regions of stemness-related genes. The characterization of this stemness-promoting BRD4-dependent regulatory pathway in SCC is of particular interest given that SCC associates with high levels of recurrence that is likely driven by the presence of CSC populations in SCC masses.
The key conclusion made by Fisher et al. (5), builds upon a considerable body of experimental evidence documenting the roles of BRD4 as a super-enhancer (SE) and P63 as a transcription factor (TF) in SC biology. BRD4 has been previously recognized as a critical regulator of the embryonic SC (ESC) transcription factors homeobox transcription factor Nanog (NANOG) (6) and OCT4 (7), which sustain ESC self-renewal and stemness properties. Additionally, several studies have suggested that ΔNp63 is indispensable for the maintenance of SC self-renewal potential in numerous normal and tumorous tissues including adult epithelia and glandular tissues (8,9) by enhancing the expression of the fibroblast growth factor receptor 2 (FGFR2) and the jagged canonical notch ligand 2 (JAG2) (10) among others. Ectopic expression of ΔNp63 in the human breast carcinoma cell line MCF-7 was reported to increase the percentage of CSCs and to enhance cancer cell clonogenicity (9) possibly by engaging the sonic hedgehog signaling pathway (8).
BRD4, through its two tandem bromodomains (BD1, BD2), binds acetylated lysine residues on H3 and H4 histones and subsequently recruits mediator complexes, the positive transcription elongation factor b (P-TEFb) and RNA polymerase II to promote the initiation and elongation steps in target gene transcription (Figure 1). Greater than 70 genes involved in normal and tumor cell survival, growth and differentiation are known to be regulated by BRD4 (11). In fact, the role of BRD4 in cancer extends beyond transcriptional and epigenetic regulation as it contributes to checkpoint activation, telomere homeostasis and DNA damage repair (11). Numerous studies have established that BRD4 is overexpressed in SCC and that its overexpression associates with poor prognosis and reduced patient survival (12). The BET inhibitor JQ1 was reported to significantly reduce the ability of BRD4 to bind H3K27 acetylation-enriched residues in the matrix metallopeptidase 2 (MMP2) gene and hence epigenetically suppress oral squamous cell carcinoma (OSCC) proliferation, migration and invasion (13). While inhibition of BRD4 in primary and transformed skin SCCs was shown to inhibit proliferation and induce apoptosis and cell cycle arrest, its overexpression reversed these effects and promoted tumor growth (14). Further investigation into the role of BRD4 in skin SCCs showed that it is required for the expression of cyclin D1, BCL-2 and MYC oncogenes (14). Similar studies have also shown that BRD4 collaborates with YAP1 (15) or JOSD1 (16) to promote the tumorigenesis of head and neck squamous cell carcinoma (HNSCC). Therefore, BRD4 is broadly recognized as an important therapeutic target in SCC; the fact that over 30 targeted inhibitors of various BET family members including BRD4 are currently in preclinical and clinical trials is testament to its perceived importance.
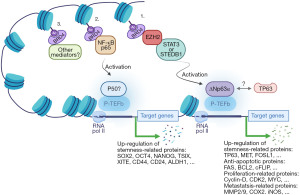
The interaction between BRD4 and EZH2 has been previously documented in different forms of SCC. Previous work has shown that BRD4 is a key upstream regulator of EZH2 in bladder cancer and that BRD4 inhibition diminishes C-MYC recruitment to the EZH2 promoter (17). This is in agreement with the Fisher group (5) observation that BRD4 regulates CSC markers via C-MYC. The ubiquitin-specific processing proteases 17 (DUB3), which is overexpressed in OSCC tissues and cell lines, was reported to promote cell proliferation and inhibit apoptosis by modulating the expression/processing of caspase-3, PARP, p21 and cyclin D1 genes (18). Interestingly, DUB3 expression in OSCC was found to positively correlate with and facilitate the production of EZH2. Further investigation into how DUB3 regulates EZH2 expression identified BRD4 as an intermediary in this process as evidenced by the observation that downregulation of DUB3 promoted BRD4 ubiquitination and that inhibition of BRD4 repressed DUB3-induced EZH2 production. EZH2, which methylates histone H3K27 along with another set domain-containing oncogenic methyltransferase (SET domain bifurcated 1; SETDB1) that methylates histone H3K9, were recently reported to play a critical role in the proliferation of HSC-5, FaDu, and Cal33 SCC cell lines (19); the same cell lines used in the Fisher et al. study (5). Both EZH2 and SETDB1 were also shown to be highly expressed in an aggressive CSC sub-population of SCC (19). Interestingly, while SETDB1 was found to regulate the SCC-derived CSC population through cooperation with ΔNp63α, the upstream EZH2 mediator activated the SETDB1/ΔNp63α pair by repressing the tumor suppressor RUNX3 (19). Another recent study by Dong et al. (4) showed that BRD4 recruits NF-κBp65 to form SEs at cancer stemness-related genes including TP63, MET and FOSL1 along with multiple mediators and oncogenic transcripts instead of MYC. The Dong et al. study also showed that disruption of SEs by the BET inhibitor JQ1 suppressed HNSCC invasive growth and metastasis by abolishing B cell-specific Moloney murine leukemia virus integration site (BMI1+) CSCs as well as proliferating non-stem tumor cells (4). It is worth noting that the BMI1 functions as a ring finger protein in the polycomb repressive complex 1 (PRC1) and that EZH2 is a functional enzymatic component of PRC2. This possibly points to the involvement of PRCs in CSCs in a way that parallels PRCs’ involvement in SC lineage commitment. Additionally, the observation that BRD4 recruits NF-κBp65 to regulate the expression of stemness-related genes including TP63 (4) and that ΔNp63α is a splice variant of P63 (20), speaks of a possible BRD4-orchestrated loop whereby NF-κBp65-induced TP63 may feed into the “EZH2-dependent stemness pathway” (Figure 1).
The role of ΔNp63α, one of the two original N-terminal splice variants of P63 (20), in SCC tumorigenesis and metastasis is well established. Overexpression of ∆Np63α, which has been documented in SCC of the lung, cervix, head and neck and esophagus, associates with poor prognosis and reduced patient survival (21). ΔNp63α has been shown to promote tumor growth, stemness, and metastasis by blocking the tumor suppressor activity of P53 family members (P53, P63 and P73) (22). ∆Np63α was reported to suppress apoptosis in SCC through its ability to interact with histone deacetylase 1 (HDAC1) and HDAC2 (23). Previous work has also shown that high ΔNp63α expression in HNSCC cell lines correlates with increased expression of the metastasis-promoting heat-shock protein 70 (HSP70) gene (24). ΔNp63α was also shown to regulate HSP70 gene transcription by recruiting additional TFs including the CCAAT binding factor in the case of HNSCC (25) and E2F1 transcription factor 1 in the case of bladder cancer (24). The involvement of ∆Np63 in the initiation and maintenance of CSCs can also be discerned by the observation that its expression is regulated by the Hedgehog and the epidermal growth factor receptor (EGFR)/STAT3 stem-cell renewal-related pathways (8) and the observation that ∆Np63α and STAT3 signaling is required for CSCs maintenance (26).
The preceding discussion points to the presence of a BRD4-EZH2-ΔNp63α axis that promotes CSC self-renewal and potency in SCCs (Figure 1) and which can be further interrogated to identify new therapeutic targets in chemoresistant and recurrent SCCs. Additionally, with the versatile transcriptional and non-transcriptional roles BRD4 plays in cell survival and growth in mind, the realization that it is a player in this stemness axis promises to unveil scores of new players in CSC biology (Figure 2). That said, there are a number of concerns that need to be considered as research work in this area progresses: (I) both the Fisher et al. study (5) and the Balinth et al. study (19), used SCC cell lines to enrich for CSCs. It is true that the Fisher et al. study showed that patient-derived SCC tumor arrays are positive for BRD4, P63, EZH2 and SOX4 but this is hardly convincing as evidence of the presence of CSCs in patient samples. For one thing, the frequency of CSCs in any cancer mass is presumably small so how can such low frequency cells account for such high levels of intensity as presented in their study. Furthermore, as shown in Figure 2, co-expression analysis of publically available SCC transcriptomic datasets showed that HNSCC, as a representative of SCC, is highly positive for these and other SC markers. Therefore, additional high-resolution evidence documenting the presence of CSCs in clinical samples of SCC need to be furnished should the observation that SCCs do indeed harbor CSCs be broadly accepted. (II) A deeper understanding of the role of the tumor microenvironment (growth requirements, immune surveillance, etc.) in the rise and preservation of CSCs is in order. (III) The potential involvement of BET members other than BRD4, mediators other than EZH2 and TFs other than ∆Np63α in inducing and/or maintaining CSCs in SCC is also worth considering. (IV) As suggested earlier (4), one is tempted to believe that SCC recurrence is at least partially precipitated by the presence of CSCs that are missed by anti-SCC drugs in current use. But this is just an assumption which still needs to be further investigated. But should this be the case, what is next? Are there any available drugs to target CSCs? Is there a need to develop new classes of drugs tailor-made for this purpose? Will there be a need to devise new patient management protocols to deal with the tumor itself and the CSC populations it harbors? Will such CSC populations prove to be homogenous or will they be heterogeneous and how will that bear on future efforts to deal with them? In short, the plot is thickening, stay tuned!
Acknowledgments
The authors wish to acknowledge the generous support of the Research Institute of Medical and Health Sciences, University of Sharjah, United Arab Emirates (UAE).
Funding: This work was supported by a University of Sharjah research grant (CoV19-0305/MH) and a Sharjah Research Academy/University of Sharjah collaborative grant (2101050170/MH), UAE.
Footnote
Provenance and Peer Review: This article was commissioned by the editorial office, Stem Cell Investigation. The article did not undergo external peer review.
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://sci.amegroups.com/article/view/10.21037/sci-2022-033/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Leiter U, Eigentler T, Garbe C. Epidemiology of skin cancer. Adv Exp Med Biol 2014;810:120-40. [PubMed]
- Apalla Z, Nashan D, Weller RB, et al. Skin Cancer: Epidemiology, Disease Burden, Pathophysiology, Diagnosis, and Therapeutic Approaches. Dermatol Ther (Heidelb) 2017;7:5-19. [Crossref] [PubMed]
- Zhang YC, Zhou Q, Wu YL. Emerging challenges of advanced squamous cell lung cancer. ESMO Open 2016;1:e000129. [PubMed]
- Dong J, Li J, Li Y, et al. Transcriptional super-enhancers control cancer stemness and metastasis genes in squamous cell carcinoma. Nat Commun 2021;12:3974. [Crossref] [PubMed]
- Fisher ML, Balinth S, Hwangbo Y, et al. BRD4 Regulates Transcription Factor ΔNp63α to Drive a Cancer Stem Cell Phenotype in Squamous Cell Carcinomas. Cancer Res 2021;81:6246-58. [Crossref] [PubMed]
- Liu W, Stein P, Cheng X, et al. BRD4 regulates Nanog expression in mouse embryonic stem cells and preimplantation embryos. Cell Death Differ 2014;21:1950-60. [Crossref] [PubMed]
- Wu T, Pinto HB, Kamikawa YF, et al. The BET family member BRD4 interacts with OCT4 and regulates pluripotency gene expression. Stem Cell Reports 2015;4:390-403. [Crossref] [PubMed]
- Memmi EM, Sanarico AG, Giacobbe A, et al. p63 Sustains self-renewal of mammary cancer stem cells through regulation of Sonic Hedgehog signaling. Proc Natl Acad Sci U S A 2015;112:3499-504. [Crossref] [PubMed]
- Du Z, Li J, Wang L, et al. Overexpression of ΔNp63α induces a stem cell phenotype in MCF7 breast carcinoma cell line through the Notch pathway. Cancer Sci 2010;101:2417-24. [Crossref] [PubMed]
- Candi E, Rufini A, Terrinoni A, et al. DeltaNp63 regulates thymic development through enhanced expression of FgfR2 and Jag2. Proc Natl Acad Sci U S A 2007;104:11999-2004. [Crossref] [PubMed]
- Donati B, Lorenzini E, Ciarrocchi A. BRD4 and Cancer: going beyond transcriptional regulation. Mol Cancer 2018;17:164. [Crossref] [PubMed]
- Gao XZ, Li WC, Diao CY, et al. Expression of BRD4 in squamous cell carcinoma and its effects on cell proliferation and invasion ability. Zhonghua Bing Li Xue Za Zhi 2018;47:344-8. [PubMed]
- Yamamoto T, Hirosue A, Nakamoto M, et al. BRD4 promotes metastatic potential in oral squamous cell carcinoma through the epigenetic regulation of the MMP2 gene. Br J Cancer 2020;123:580-90. [Crossref] [PubMed]
- Xiang T, Bai JY, She C, et al. Bromodomain protein BRD4 promotes cell proliferation in skin squamous cell carcinoma. Cell Signal 2018;42:106-13. [Crossref] [PubMed]
- Chen N, Golczer G, Ghose S, et al. YAP1 maintains active chromatin state in head and neck squamous cell carcinomas that promotes tumorigenesis through cooperation with BRD4. Cell Rep 2022;39:110970. [Crossref] [PubMed]
- Jing C, Liu D, Lai Q, et al. JOSD1 promotes proliferation and chemoresistance of head and neck squamous cell carcinoma under the epigenetic regulation of BRD4. Cancer Cell Int 2021;21:375. [Crossref] [PubMed]
- Wu X, Liu D, Tao D, et al. BRD4 Regulates EZH2 Transcription through Upregulation of C-MYC and Represents a Novel Therapeutic Target in Bladder Cancer. Mol Cancer Ther 2016;15:1029-42. [Crossref] [PubMed]
- Luo F, Zhou Z, Cai J, et al. DUB3 Facilitates Growth and Inhibits Apoptosis Through Enhancing Expression of EZH2 in Oral Squamous Cell Carcinoma. Onco Targets Ther 2020;13:1447-60. [Crossref] [PubMed]
- Balinth S, Fisher ML, Hwangbo Y, et al. EZH2 regulates a SETDB1/ΔNp63α axis via RUNX3 to drive a cancer stem cell phenotype in squamous cell carcinoma. Oncogene 2022;41:4130-44. [Crossref] [PubMed]
- Mangiulli M, Valletti A, Caratozzolo MF, et al. Identification and functional characterization of two new transcriptional variants of the human p63 gene. Nucleic Acids Res 2009;37:6092-104. [Crossref] [PubMed]
- Gatti V, Fierro C, Annicchiarico-Petruzzelli M, et al. ΔNp63 in squamous cell carcinoma: defining the oncogenic routes affecting epigenetic landscape and tumour microenvironment. Mol Oncol 2019;13:981-1001. [Crossref] [PubMed]
- Nekulova M, Holcakova J, Coates P, et al. The role of p63 in cancer, stem cells and cancer stem cells. Cell Mol Biol Lett 2011;16:296-327. [Crossref] [PubMed]
- Ramsey MR, He L, Forster N, et al. Physical association of HDAC1 and HDAC2 with p63 mediates transcriptional repression and tumor maintenance in squamous cell carcinoma. Cancer Res 2011;71:4373-9. [Crossref] [PubMed]
- Jin H, Xie Q, Guo X, et al. p63α protein up-regulates heat shock protein 70 expression via E2F1 transcription factor 1, promoting Wasf3/Wave3/MMP9 signaling and bladder cancer invasion. J Biol Chem 2017;292:15952-63. [Crossref] [PubMed]
- Wu G, Osada M, Guo Z, et al. DeltaNp63alpha up-regulates the Hsp70 gene in human cancer. Cancer Res 2005;65:758-66. [Crossref] [PubMed]
- Wei S, Li J, Tang M, et al. STAT3 and p63 in the Regulation of Cancer Stemness. Front Genet 2022;13:909251. [Crossref] [PubMed]
Cite this article as: Hamad M, Ali A, Muhammad JS. BRD4 regulates the induction and maintenance of cancer stem cells in squamous cell carcinoma. Stem Cell Investig 2022;9:6.


