
Optimization of adeno-associated virus (AAV) gene delivery into human bone marrow stem cells (hBMSCs)
Introduction
Efficient nucleic acid delivery systems are vital for studying gene function and performing gene therapies. Physical, chemical and biological-based methods are commonly used to deliver genes into mammalian cells. Physical methods (electroporation, sonoporation, microinjection and biolistics or bombardment, etc.) use physical forces to create temporary membrane pores to allow entry of the nucleic acid into cells (1). However, such methods require expensive equipment and often are inconvenient for most gene delivery applications (2). More importantly, physical gene delivery generally exhibits lower cell viability due to heavy trauma and apoptotic or programmed cell death caused or induced by the physical forces, thus having limited applications (3). Chemical methods use natural or synthetic compounds, namely transfection reagents, as nucleic acid carrier molecules to facilitate nucleic acid delivery into cells. Transfection reagents are classified into four major types: calcium phosphate, DEAE-dextran, cationic lipid, and cationic polymer, and each has advantages and disadvantages (4,5). Cell toxicity and low transfection efficiency for large DNA (plasmids) are the main disadvantages of the chemical methods. Biological methods rely on the infectious property of bacteria or viruses to transfer genes into cells. Viral gene delivery is the most studied method for delivering transgenes into animal cells. If appropriately optimized, it can be the most efficient gene delivery method. Adenoviruses, retroviruses, lentiviruses, and adeno-associated viruses (AAVs) have been used to transfer genes in vitro and in vivo. Of them, AAVs are attractive virus-based gene delivery candidates as they are innately nonpathogenic and cause a very mild immune response, unlike adenoviruses and retroviruses (including lentiviruses), which can cause severe immunogenicity and other problems (6). For example, genes delivered by retroviruses (or lentiviruses) are randomly inserted into the genome of the host cells, causing mutations and other safety issues (such as tumorigenesis). Genes delivered by AAV generally do not integrate into the host genome or integrate with very low frequency in a site-specific manner (7).
Mesenchymal stem cells (MSCs) are adult stem cells primarily residing in the bone marrow. Human bone marrow stem cells (hBMSCs) possess the multipotent capability to differentiate into osteoblasts, adipocytes, chondrocytes, etc. (8) and are the most frequently used stem cells in cell therapy and tissue engineering (9). Directing hBMSC differentiation toward desired cell types is vital for the applications of the cells in tissue engineering and regenerative medicine. However, the molecular pathways regulating their differentiation are not fully understood. Developing highly efficient gene delivery methods is essential for molecular biology experiments to elucidate the genes controlling hBMSC differentiation for cell- and gene-based therapies. In this regard, gene delivery into hBMSCs using AAV vectors has been attempted. Ju et al. and Ito et al. first reported hBMSC transduction with AAV (10,11). However, subsequent studies in different laboratories reported inconsistent transduction efficiency of AAVs for hBMSCs showing a high variation of transduction for AAV vectors in human MSCs (12). The source of variation in AAV vector transduction is currently unknown, likely due to multiple factors, such as, cell culture condition, vector preparation, and transduction conditions (12). A publication indicated that AAV transduction efficiencies could achieve up to 65% (13). Thus, further improving AAV transduction is desired. The objectives of this study were to further optimize the AAV transduction and attempt to establish a highly efficient AAV gene delivery system for the in vitro transduction of hBMSCs. Additionally, it is our goal to study the longevity of transgene expression in hBMSCs as long-term transgene expression in human MSCs has yet to be conclusively demonstrated after AAV transduction.
So far, 13 AAV serotypes (with variants within serotypes) have been identified with variable tissue and cell type tropism. It is critical to test different AAVs to achieve optimal gene delivery for various cells and tissues. We selected the six AAV variants for this project for the following reasons: (I) AAV2 was selected because it has been reported to deliver genes into hBMSCs successfully; (II) AAV1 and AAV6 were selected because a study showed they have a great ability to transduce a wide range of cell types (14); (III) AAV-DJ is an engineered chimeric capsid of AAV-2, 8 and 9 containing a heparin-binding domain in its capsid, which may efficiently transduce a broad range of cell types and escape immune neutralization (15). It was worth testing this serotype on hBMSCs. Therefore, AAV-DJ was selected for our initial experiment. AAV2.7m8 is also a synthetic AAV containing a 10-amino acid peptide inserted into the AAV2 capsid protein involved in viral binding to the primary receptor (16).
Methods
Culture of hBMSCs
Primary hBMSCs (Passage 0) isolated from two donors, which are termed hBMSCs-1 and hBMSCs-2 in this study, were purchased from Obatala Sciences (New Orleans, LA, USA). The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Purchasing and using human stem cells for research have been approved by the IRB Office of Louisiana State University (IRB office ID: IORG0000106). Written informed consent is not required as no patients involved in this study. The cells were cultured in T-75 flasks with a medium consisting of MEM-α (Corning) plus 20% FBS (Neuromics) and 1% Penicillin-Streptomycin (Gibco) in 5% CO2 at 37 °C. The culture medium was changed every four days. Cells were passaged at 80–90% confluence at a 1:3 ratio, and different passages were obtained and cryopreserved in liquid N2. Passages 4 and 5 cells were used for the experiments.
AAVs and transduction experiments
AAVs containing enhanced green fluorescence protein (eGFP) under transcriptional control by a CMV promoter (i.e., AAVs-eGFP) were purchased from VectorBuilder (Chicago, IL, USA). The AAV titers provided by the manufacturer were based on qPCR assessment. The titer of the AAV stocks was adjusted to 1012 GC/mL with phosphate-buffered saline (PBS) buffer supplemented with 200 mM NaCl and 0.00067% pluronic F-127. For assessment of transduction efficiency, hBMSCs were seeded in 24-well plates with cell growth medium (MEM-α plus 20% FBS and 1% Penicillin-Streptomycin) in 5% CO2 at 37 °C. When the cells grew to about 90% confluency, AAVs-eGFP were added to 200 µL of the growth medium in each well with the designated multiplicity of infection (MOI) for infection. The infection medium was replaced with a fresh cell growth medium (0.5 mL/well) after overnight incubation, and the cells were cultured at normal cell culture conditions. To assess the mRNA (transcription) of the eGFP, we seeded cells in 12-well plates for AAV2-eGFP infection as described above with 0.5 mL medium/well. Infected cells were cultured in cell growth medium (1 mL/well) with four days of medium change interval and collected at designated time points for RNA extraction.
Assessment of transduction efficiency
Hoechst 33324 was used to stain the cell nuclei to facilitate counting the cell numbers. Briefly, cells were stained with Hoechst 33324 (5 µg/mL) in cell growth medium for 5 min on days 5, 10, and 15 post-infections. The staining medium was removed, and the cells were washed once with a fresh cell growth medium (0.5 mL/well) to remove the Hoechst dye residual. Three distinct microscopic fields were visualized and photographed first for blue-fluorescent nuclei and then the green-fluorescent image of the same view using a ZOETM Fluorescent Cell Imager (Bio-Rad). The images were analyzed with ImageJ to acquire total cell numbers by counting blue-fluorescent nuclei. The blue-fluorescent images were merged with the corresponding green-fluorescent images, and green-fluorescent cells (i.e., eGFP-positive cells) were counted twice manually to ensure accuracy. Transduction efficiency was expressed by the percentage of green-fluorescent cells over the total cells.
Assessment of longevity of eGFP mRNA expression post-AAV infection
hBMSCs were seeded in 12-well plates and incubated with 20K MOI of AAV2-eGFP. Cells were collected in TRI reagent (Molecular Research Center, Cincinnati, OH, USA) on days 5, 10, 15, 20, 25, and 30 post-infection for total RNA extraction using the traditional TRIzol method (Molecular Research Center, Inc., Cincinnati, OH, USA). Extracted RNA was digested with TurboTM DNase (ThermoFisher Scientific) to remove possible DNA contamination, followed by quantitation by NanoDropTM 8000 Spectrophotometer (ThermoFisher Scientific). The RNA quality was assessed by OD260/280 and OD260/230; 700–900 ng total RNA was reverse transcribed to 20 µL cDNA with random primers and M-MLV reverse transcriptase (ThermoFisher Scientific). Next, conventional PCR of 25 cycles was performed by mixing 1 µL cDNA, 300 nM primers (Table 1) with Maxime™ PCR PreMix (i-StarTaq) (iNtRON Biotechnology) at 20 µL reaction volume to detect eGFP mRNA expression. The PCR product was loaded to 1.5% agarose gel containing ethidium bromide for electrophoresis, followed by gel imaging with Bio-Rad ChemiDoc + gel imaging system. For qPCR analysis, 1 µL cDNA was mixed with iTaq Universal SYBR Green Supermix (Bio-Rad) and primers (Table 1) at 20 µL. CT values were acquired by running the reactions with CFX Opus 96 Real-Time PCR Instrument (Bio-Rad).
Table 1
| Gene | Forward primer sequence 5'—3' | Reverse primer sequence 5'—3' | Note |
|---|---|---|---|
| eGFP | CACATGAAGCAGCACGACTTC | GACTGGGTGCTCAGGTAGTG | For PCR |
| eGFP | CAAGATCCGCCACAACATCG | GACTGGGTGCTCAGGTAGTG | For qPCR |
| Beta-Actin | CCACCATGTACCCTGGCATT | TGTGCAATCAAAGTCCTCGG | For PCR |
| Beta-Actin | TCGTGCGTGACATTAAGGAG | GTCAGGCAGCTCGTAGCTCT | For qPCR |
PCR, polymerase chain reaction; qPCR, quantitative polymerase chain reaction.
Statistical analysis
Statistical analysis was performed with the Wilcoxon rank-sum test or ANOVA flowed by Tukey’s test using SAS program. The P values equal or less than 0.05 (*), 0.01 (**) and 0.001 (***) indicate that the differences between means are statistically significant or highly statistical significance, respectively. Data were presented as mean ± standard deviation (SD).
Results
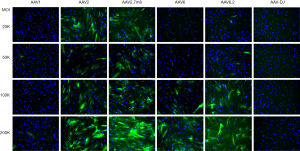
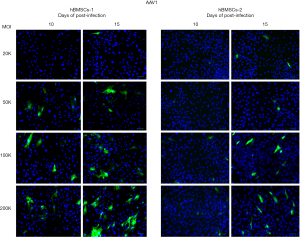
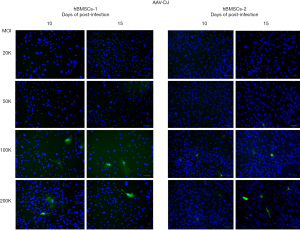
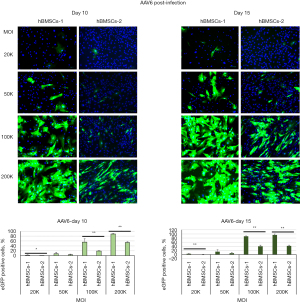
Weak green fluorescence started to appear as early as three days post-infection for AAV2, AAV2.7m8, AAV6, and AAV6.2 in hBMSCs-1 and hBMSCs-2. And the green fluorescence became bright on day 5 post-infection in some cells, especially for AAV2 and AAV2.7m8 at the high MOI treatments (Figure 1). No green fluorescence could be observed in cells infected with AAV1 and AAV-DJ on day 5 (Figure 1). Only a few green-fluorescent cells (i.e., eGFP+ cells) were seen in AAV1 and AAV-DJ infected cells on days 10 and 15 post-infection (Figures 2,3). AAV1 treatment showed slightly more green-fluorescent cells than AAV-DJ treatment on days 10 and 15 post-infection (Figures 2,3) in both cell lines. The results indicated that AAV1 and AAV-DJ were very inefficient for the transduction of hBMSCs. Thus, AAV1 and AAV-DJ were excluded for further analyses and optimization for hBMSCs transduction. eGFP-positive cells were significantly increased for other serotypes on days 10 and 15. We counted total cells and green-fluorescent cells and calculated the percentage of eGFP-positive cells. The results are shown in Figures 4-7.
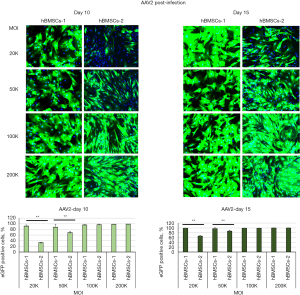
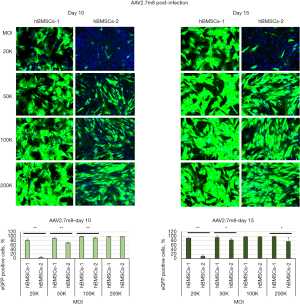
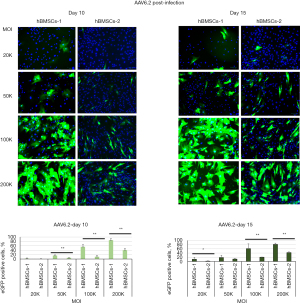
For AAV2-infected cells, more eGFP-positive cells could be seen in hBMSCs-1 than in hBMSCs-2 at 20K and 50K MOI on days 10 and 15 (Figure 4). For example, more than 90% of the AAV2-treated cells were eGFP positive in hBMSCs-1, but less than 40% of cells were eGFP positive in hBMSCs-2 at 20K MOI on day 10 post-infection. The difference was highly statistically significant (P≤0.01). However, at MOI of 100K and 200K, almost all cells in hBMSCs-1 and hBMSCs-2 exhibited eGFP positive (Figure 4), and there was no significant difference. AAV2.7m8 showed a similar trend as AAV2 (Figure 5). At MOI of 20K, very few eGFP-positive cells were seen in hBMSCs-2, whereas most of the cells in hBMSCs-1 were eGFP positive. However, when MOI was greater than 100K, most cells (>80%) in both hBMSCs-1 and hBMSCs-2 were eGFP positive, although hBMSCs-1 still had higher transduction efficiency than hBMSCs-2.
MOIs and the donor (genetic) background of the hBMSCs appeared to have a huge impact on transduction efficiency for AAV6 and AAV6.2. Very few eGFP-positive cells (less than 10%) were seen in the treatments of MOIs 20K and 50K. Significant increases in eGFP-positive cells were observed as MOI increased to 100K and 200K. Approximately 80–90% of cells were eGFP positive for hBMSCs-1, and 40% were eGFP positive for hBMSCs-2 on day 15 post-infection. The differences in the percentage of eGFP-positive cells between hBMSCs-1 and hBMSCs-2 were statistically significant (Figures 6,7).
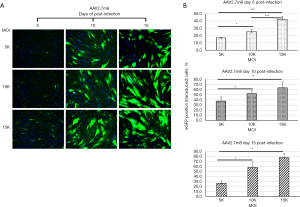
The above results made it clear that AAV2 and AAV2.7m8 were superior to other AAV serotypes for the transduction of hBMSCs. The transduction efficiency of AAV2 and AAV2.7m8 was less affected by the MOIs, and cell donor backgrounds compared to AAV6 and AAV6.2. Since it is desired to use as low as possible MOI of AAVs to infect cells for gene delivery, we reasoned it is possible to reduce the MOI of AAV2 and AAV2.7m8. Therefore, we tested AAV2 and AAV2.7m8 at reduced MOIs of 5K, 10K, and 15K for infection of hBMSCs-1. The results are presented in Figures 8,9. Greater than 80% of cells could be transduced at MOIs of 10K and 15K in AAV2-infected cells on day 15 of post-infection (Figure 8). AAV2.7m8 also showed about 80% transduction with an MOI of 15K on day 15 (Figure 9).
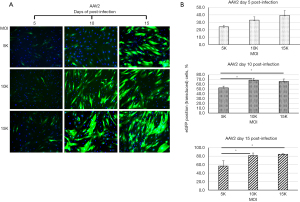
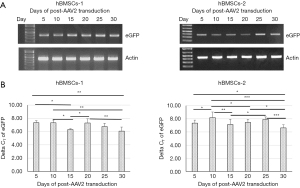
The longevity of transgene expression is a critical parameter for assessing gene delivery systems in gene therapy and gene functional studies. Thus, we collected AAV2-eGFP-infected hBMSCs at different time points for conventional and quantitative RT-PCR analysis. qRT-PCR data was reported as delta CT by normalizing eGFP CT to beta-Actin CT (i.e., delta CT = eGFP CT − Actin CT). The results indicated that the transcription (mRNA) of the transgene (eGFP) could be detected on days 5 to 30 post-AAV2-infection (Figure 10). However, the qRT-PCR analysis showed increased expression of eGFP appeared after day 15 post-infection (Figure 10B). however, there was a variation between hBMSCs-1 and hBMSCs-2. The delta CT of hBMSCs-1 was generally 0.5–1 cycles lower than the delta CT of hBMSCs-2 for most days, indicating hBMSCs-1 expressed higher levels of eGFP mRNA (Figure 10B). On average, maximal eGFP mRNA levels were seen on day 30 post-infection for both cell lines (Figure 10B).
Discussion
AAV vectors have been extensively studied for their therapeutic applications and biomedical research. The molecular mechanism of AAV infection and cell entry is still largely unknown (17). There are many serotypes and variants of AAVs, and different AAV serotypes display significant differences in transduction efficiency and cell and tissue tropism (18,19). The selection of appropriate AAV serotypes is among the first things that need to do in designing AAV gene delivery. Here, we tested selected serotypes for delivering the eGFP reporter gene into hBMSCs derived from different donors. Our data indicate that serotypes are the most critical factor. Based on our observation, we rank the superiority of those AAVs for transducing hBMSCs as follows regardless of cell donor background: AAV2 > AAV2.7m8 > AAV6 > AAV6.2 > AAV1 > AAV-DJ.
Using AAVs to deliver transgenes into human MSCs has been attempted, and published data regarding the AAV vector transduction efficiency of stem cells are inconsistent (12). An early study compared AAV serotypes 1, 2, 3, 4, 5, 6, and 8 to infect MSCs of human and non-human primates and found that AAV2 was the most efficient serotype for human and baboon MSCs (20). Another study reported that up to 65% transduction could be achieved with AAV2 in human MSCs at 4 days post-transduction (13). Our data align with those published results showing that AAV2 is the most efficient serotype for transducing hBMSCs. Our experiments showed AAV2 is highly efficient in transferring genes into hBMSCs. High efficiency of transduction (>80%) could be achieved by simply adding the viral particles to the cell cultures at MOI as low as 10K. Transduction efficiency could reach almost 100% when hBMSCs were infected with high MOI (≥100K) of AAV2 or its variant AAV2.7m8. However, our results suggested that the cell donor’s genetic background could affect the AAV transducing efficiency, especially at the lower MOIs. We noticed that the hBMSCs derived from different donors had a significant impact on transduction efficiency at a low MOI. For example, hBMSCs derived from donor 1 seem to result in higher transduction efficiency than the hBMSCs from donor 2; suggesting that hBMSCs’ genetic background may impact the transduction efficiency.
It is worth noting that although green fluorescence could be observed on day 5 post-AAV infection, as shown in Figure 1, it would be a better time to assess the transduction efficiency on post-infection days 10–15. The reason is as follows: GFP is a highly stable protein with a reported half-life of 26–54 hours in most cells (21-23). The fluorescence of eGFP is greater than GFP in human cells (24). Thus, cells could accumulate high-level GFP or eGFP protein over time such that an increase in fluorescence could be observed (Figures 4,5). However, the high stability of GFP or eGFP makes it unsuitable for real-time evaluate its transcription based on the green-fluorescent intensity. Thus, we assessed the eGFP transcription on different post-AAV2 infection days by the mRNA using RT-PCR and qRT-PCR (Figure 10), as the eGFP mRNA half-life was reported to be 4.8 hours in human cells (24), thus better reflect the real-time eGFP transcription.
In a recent publication, Bougioukli et al. reported that AAV2 and AAV6 had limited potential for delivering transgenes (eGFP and BMP-2) into hBMSCs on day 7 post-infection (25). Our experiment showed no or very weak green fluorescence in cells infected with AAV6-eGFP below 100K MOI on day 5 post-infection (Figure 1). However, green fluorescence was seen in the cells infected with AAV2-eGFP of 5K to 200K MOIs on day 5 post-infection (Figures 1,8). We observed the transgene (eGFP) expression on day 5 post-AAV2-eGFP infection; however, it appeared that the increased expression was not reached until after day 15 post-infection based on qRT-PCR analysis. AAV has a single-stranded DNA (ssDNA) genome (i.e., ssAAV). After entering the cells, the ssDNA must be converted to double-stranded DNA (dsDNA) before the transgene can be transcribed to mRNA. Thus, there is a delay in the onset of transgene expression after an AAV infection. The ssDNA to dsDNA conversion is a well-documented, rate-limiting step involving the de novo synthesizing of the second strand DNA (26,27). We observed a dramatic increase in green fluorescence beginning on day 10 post-infection. qRT-PCR analysis indicated that the increased eGFP transcription likely occurs after day 15 post-infection, depending on cell lines. The delay of transgene expression is one of the disadvantages or limitations of using AAV vectors for gene delivery applications, especially when immediate therapeutic intervention is needed. One approach is to use self-complementary AAVs (scAAVs), which circumvent second-strand DNA synthesis requirements to overcome this limitation. scAAV has been demonstrated to exhibit faster, stronger, and prolonged transgene expression (28). However, the cargo capacity of scAAV (5’ inverted terminal repeat to 3’ inverted terminal repeat) that can be properly packaged into mature viral particles is only about half that of conventional ssAAV.
Conclusions
In summary, our data suggested that AAV1 and AAV-DJ have minimal transduction ability for hBMSCs. Although AAV6 and its variant AAV6.2 can deliver transgenes into hBMSCs, their transduction efficiency is largely affected by MOI. The use of high MOI is needed to result in considerable transduction for AAV6 and AAV6.2. In contrast, AAV2 and its variant AAV2.7m8 can efficiently deliver transgene into hBMSCs with almost 100% transduction efficiency; however, AAV2 appears superior to AAV2.7m8. At an MOI less than 50K, hBMSCs derived from different donors may exhibit a discrepancy in transduction efficiency with AAV2. However, at an MOI greater than 100K, AAV2 appears to transduce hBMSCs derived from different donors without significant difference.
Acknowledgments
The authors thank Dr. Xue Wen for her assistance in the collection and statistical analysis of the data.
Funding: This research was supported by the National Institutes of Health/National Institute of Arthritis and Musculoskeletal and Skin Diseases (1R21AR076583-01A1).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://sci.amegroups.com/article/view/10.21037/sci-2022-042/coif). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. The study was conducted in accordance with the Declaration of Helsinki (as revised in 2013). Purchasing and using human stem cells for research have been approved by the IRB Office of Louisiana State University (IRB number: E11515). Written informed consent is not required as no patients involved in this study.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Mehier-Humbert S, Guy RH. Physical methods for gene transfer: improving the kinetics of gene delivery into cells. Adv Drug Deliv Rev 2005;57:733-53. [Crossref] [PubMed]
- Bono N, Ponti F, Mantovani D, et al. Non-Viral in Vitro Gene Delivery: It is Now Time to Set the Bar! Pharmaceutics 2020;12:183. [Crossref] [PubMed]
- Mellott AJ, Forrest ML, Detamore MS. Physical non-viral gene delivery methods for tissue engineering. Ann Biomed Eng 2013;41:446-68. [Crossref] [PubMed]
- Gao X, Kim KS, Liu D. Nonviral gene delivery: what we know and what is next. AAPS J 2007;9:E92-104. [Crossref] [PubMed]
- Fus-Kujawa A, Prus P, Bajdak-Rusinek K, et al. An Overview of Methods and Tools for Transfection of Eukaryotic Cells in vitro. Front Bioeng Biotechnol 2021;9:701031. [Crossref] [PubMed]
- Samulski RJ, Muzyczka N. AAV-Mediated Gene Therapy for Research and Therapeutic Purposes. Annu Rev Virol 2014;1:427-51. [Crossref] [PubMed]
- Hamilton H, Gomos J, Berns KI, et al. Adeno-associated virus site-specific integration and AAVS1 disruption. J Virol 2004;78:7874-82. [Crossref] [PubMed]
- Kassem M, Abdallah BM. Human bone-marrow-derived mesenchymal stem cells: biological characteristics and potential role in therapy of degenerative diseases. Cell Tissue Res 2008;331:157-63. [Crossref] [PubMed]
- Fu X, Liu G, Halim A, et al. Mesenchymal Stem Cell Migration and Tissue Repair. Cells 2019;8:784. [Crossref] [PubMed]
- Ju XD, Lou SQ, Wang WG, et al. Effect of hydroxyurea and etoposide on transduction of human bone marrow mesenchymal stem and progenitor cell by adeno-associated virus vectors. Acta Pharmacol Sin 2004;25:196-202. [PubMed]
- Ito H, Goater JJ, Tiyapatanaputi P, et al. Light-activated gene transduction of recombinant adeno-associated virus in human mesenchymal stem cells. Gene Ther 2004;11:34-41. [Crossref] [PubMed]
- Brown N, Song L, Kollu NR, et al. Adeno-Associated Virus Vectors and Stem Cells: Friends or Foes? Hum Gene Ther 2017;28:450-63. [Crossref] [PubMed]
- Stender S, Murphy M, O'Brien T, et al. Adeno-associated viral vector transduction of human mesenchymal stem cells. Eur Cell Mater 2007;13:93-9; discussion 99. [Crossref] [PubMed]
- Ellis BL, Hirsch ML, Barker JC, et al. A survey of ex vivo/in vitro transduction efficiency of mammalian primary cells and cell lines with Nine natural adeno-associated virus (AAV1-9) and one engineered adeno-associated virus serotype. Virol J 2013;10:74. [Crossref] [PubMed]
- Grimm D, Lee JS, Wang L, et al. In vitro and in vivo gene therapy vector evolution via multispecies interbreeding and retargeting of adeno-associated viruses. J Virol 2008;82:5887-911. [Crossref] [PubMed]
- Khabou H, Desrosiers M, Winckler C, et al. Insight into the mechanisms of enhanced retinal transduction by the engineered AAV2 capsid variant -7m8. Biotechnol Bioeng 2016;113:2712-24. [Crossref] [PubMed]
- Berry GE, Asokan A. Cellular transduction mechanisms of adeno-associated viral vectors. Curr Opin Virol 2016;21:54-60. [Crossref] [PubMed]
- Wiley LA, Burnight ER, Kaalberg EE, et al. Assessment of Adeno-Associated Virus Serotype Tropism in Human Retinal Explants. Hum Gene Ther 2018;29:424-36. [Crossref] [PubMed]
- Srivastava A. In vivo tissue-tropism of adeno-associated viral vectors. Curr Opin Virol 2016;21:75-80. [Crossref] [PubMed]
- Chng K, Larsen SR, Zhou S, et al. Specific adeno-associated virus serotypes facilitate efficient gene transfer into human and non-human primate mesenchymal stromal cells. J Gene Med 2007;9:22-32. [Crossref] [PubMed]
- Corish P, Tyler-Smith C. Attenuation of green fluorescent protein half-life in mammalian cells. Protein Eng 1999;12:1035-40. [Crossref] [PubMed]
- Li X, Zhao X, Fang Y, et al. Generation of destabilized green fluorescent protein as a transcription reporter. J Biol Chem 1998;273:34970-5. [Crossref] [PubMed]
- Sacchetti A, El Sewedy T, Nasr AF, et al. Efficient GFP mutations profoundly affect mRNA transcription and translation rates. FEBS Lett 2001;492:151-5. [Crossref] [PubMed]
- Kudla G, Lipinski L, Caffin F, et al. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol 2006;4:e180. [Crossref] [PubMed]
- Bougioukli S, Chateau M, Morales H, et al. Limited potential of AAV-mediated gene therapy in transducing human mesenchymal stem cells for bone repair applications. Gene Ther 2021;28:729-39. [Crossref] [PubMed]
- Ferrari FK, Samulski T, Shenk T, et al. Second-strand synthesis is a rate-limiting step for efficient transduction by recombinant adeno-associated virus vectors. J Virol 1996;70:3227-34. [Crossref] [PubMed]
- Fisher KJ, Gao GP, Weitzman MD, et al. Transduction with recombinant adeno-associated virus for gene therapy is limited by leading-strand synthesis. J Virol 1996;70:520-32. [Crossref] [PubMed]
- Wang Z, Ma HI, Li J, et al. Rapid and highly efficient transduction by double-stranded adeno-associated virus vectors in vitro and in vivo. Gene Ther 2003;10:2105-11. [Crossref] [PubMed]
Cite this article as: Yao S, Rong W, Yuan Y. Optimization of adeno-associated virus (AAV) gene delivery into human bone marrow stem cells (hBMSCs). Stem Cell Investig 2023;10:3.


