
Identification and characterization of enhancer elements controlling cell type-specific and signalling dependent chromatin programming during hematopoietic development
Introduction
Embryonic development is tightly controlled at the level of gene expression. The precise control of tissue-specificity of gene expression is essential for successful development and depends on distal cis-regulatory elements such as enhancers which interact with promoter elements in physical space. The activity of these elements is controlled by transcription factors (TFs) which bind to DNA wrapped into chromatin, leading to the modification of the chromatin landscape, and the assembly of the transcription machinery. Transcription has been shown to occur within defined chromosomal locations termed topologically-associated domains (TADs) [reviewed in (1)] where TF complexes bring together control elements across large distances within the genome [reviewed in (2)].
In the developing embryo, TFs regulating the assembly/disassembly of transcriptional complexes and ultimately gene expression, are directed by complex extrinsic signalling processes which connect all cells within a multi-cellular organism to their environment. Cell-to-cell signalling is induced by specific ligands such as growth factors which activate their cognate receptor molecules. Upon binding of their respective ligands and activation, intracellular signalling cascades, often involving phophorylation are induced which eventually terminate at inducible TFs and regulate their activity. The regulation of cell growth and differentiation therefore involves the precise and coordinated interplay of cell extrinsic and intrinsic processes.
For many decades the development of the hematopoietic system has been used as a model for studying the molecular basis of cell fate decisions and gene regulation, and as such it is one of the best understood developmental pathways. In vertebrates, embryonic hematopoiesis is the process which generates hematopoietic stem cells (HSCs). These cells sit at the top of the hematopoietic hierarchy and have the ability to self-renew and give rise to all mature blood cell types in the adult organism (3). Furthermore, HSCs are maintained for life and replenish components of the blood system (4). Operationally, HSCs are defined as cells that provide long-term reconstitution of the entire hematopoietic system of an irradiated adult recipient (5).
An experimental model that has yielded important insights into the molecular details of hematopoietic specification is the differentiation of embryonic stem cells (ESCs) into blood (6,7). ESCs are derived from the inner cell mass (ICM) of the blastocyst (8-10). However, so far blood progenitor cells produced in such a system were unable to yield long-term hematopoietic reconstitution. The precise signals that control the formation of these cells and their correct gene expression patterns have been largely elusive. Understanding how signalling and the cellular environment direct the differentiation of ESCs to HSCs is therefore of great importance, as the ability to produce large quantities of HSCs capable of giving rise to any of the constituents of blood in vitro would be of significant therapeutic and biotechnological value [reviewed in (11,12)]. To achieve this aim, we need to know how HSC identity is established at the gene expression control level. We need to know the genomic location of cis-regulatory elements directing cell-type-specific control during the whole differentiation pathway, the TFs regulating their activity and which signalling pathways these TFs respond to. This review will cover (I) our current understanding of embryonic hematopoiesis, (II) the use of in vitro differentiation systems for study and production of HSCs and (III) the methods used for the identification and study of signalling responsive cis-elements and the TFs regulating these processes.
Embryonic hematopoietic development occurs in several waves
Pluripotent cells from the blastocyst give rise to any of the three germ layers: endoderm, mesoderm and ectoderm (13,14). In the mouse embryo primary germ-layer specification occurs between embryonic days (E) 4.5 and E7.5 (15). Hematopoietic specification occurs from the mesodermal germ layer in three waves. The first wave, known as ‘primitive hematopoiesis’, occurs at around E7 in the blood islands of the yolk sack producing mature primitive erythrocytes, macrophages and megakaryocytes (16-18). This wave does not produce HSCs, instead, it provides short-lasting hematopoietic cells required for the embryo’s needs, such as oxygen supply, tissue remodelling and vascular maintenance (19,20). At E8.25 the second hematopoietic wave produces both erythroid-myeloid progenitors (EMPs) and long lasting late EMPs which can differentiate into cells displaying adult blood cell characteristics and functions (21,22). At this timepoint, embryonic lymphoid commitment begins through the emergence of immune-restricted and lymphoid-primed progenitors which contributes to the establishment of lymphoid and myeloid components of the immune system (23). However, no HSCs that fulfil the criteria above are produced.
In mice the final wave of blood cell development occurs at E10.5 [E27 to E40 in humans (24)] and gives rise to the definitive HSCs which emerge from the ventral section of the dorsal aorta in a region termed the aorta-gonad-mesonephros (AGM) which is derived from the mesodermal germ layer (5,25). It is these cells which are capable of reconstituting the entire adult hematopoietic system. In response to paracrine and autocrine signalling HSCs develop from a specialised hemogenic endothelium (HE) which overlays the dorsal mesenchyme within the AGM. The HE then undergoes an endothelial-to-hematopoietic transition (EHT) (19,26-29) during which flat endothelial cells buldge upwards towards the intra-aortic lumen (30-32) by forming clusters, lose their endothelial transcriptomic signature and adopt a hematopoietic phenotype (33). Once formed and after maturation (34), HSCs bud off, enter the bloodstream and colonise the foetal liver and then subsequently the bone marrow where they generate all hematopoietic cell types (35). However, the dorsal aorta is not the only endothelial layer capable of generating blood cells, also the earlier waves of blood cell formation are formed by an EHT process (36).
Hematopoietic development from ESCs is regulated by dynamic gene regulatory networks (GRNs)
TFs and their respective targets, including genes that encode TFs themselves, form GRNs which define the identity of a cell (37,38). It therefore follows that in development, different cellular identities are established by changes from one GRN to another. To understand this process in molecular detail, it is necessary to (I) identify cell type-specific cis-regulatory elements, (II) how these impact on gene expression, (III) identify the transcriptional complexes which bind to them and (IV) to understand how these respond to external cues.
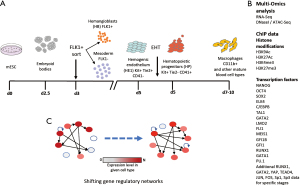
The in vitro differentiation of ESCs into blood cells which is representative of the second wave of blood cell development has proven instrumental in gaining a mechanistic understanding of this process. Goode et al. (39) employed a culture system in which mouse ES cells are replated into a culture medium that does not support pluripotency. Replating leads to the formation of embryoid bodies within which hematopoietic specification takes place, and from which the different cell types from this pathway can be isolated by employing cell sorting (Figure 1A). Cell types include mesodermal cells and cells expressing the endothelial marker FLK1 (the receptor for the endothelial growth factor VEGFA encoded by the Kdr gene). These cells, which give rise to both endothelial and hematopoietic cells, are purified using cell sorting and replated in a culture medium that supports the formation of the hemogenic endothelium (HE1). From these cells, the first steps of hematopoietic commitment take place with the down-regulation of the endothelial marker Tie2 and the up-regulation of the CD41 marker, generating HE2 cells which are still adherent. Thereafter, cells bud off and form hematopoietic progenitor (HP) cells through the EHT. When exposed to the right cytokines, HP cells are capable of forming both lymphoid and myeloid cells, exemplified here by macrophages (Figure 1A). In order to examine, how the interplay of TFs and chromatin components drives gene expression at the different stages of blood cell specification, Goode et al. (39) generated global multi-omics data on measuring TF binding, gene expression, histone modifications and open chromatin regions (listed in Figure 1B) during six sequential stages of hematopoietic differentiation from mouse ESCs to macrophages, including HE and HP. The work revealed the chromatin signature of potential regulatory elements and the TF binding patterns driving the differential gene expression required for hematopoietic lineage commitment. It allowed to group chromatin states into “inactive” (H3K27me3), “poised” (“H3K4me3/H3K27me3), “active” (H3K4me3 or H3K27Ac) and “unmarked”. In addition, integrating TF binding, gene expression and chromatin structure, this work allowed to construct a comprehensive dynamic core regulatory network model for hematopoietic specification with network connections being rewired during differentiation (represented in Figure 1C). Moreover, beyond what could be measured by chromatin immunoprecipitation (ChIP) assays the work highlighted the relative importance of TFs with respect to their importance for the maintenance of specific cell types which is encoded in the binding motif composition of cell-type specific cis-regulatory elements (44). The analysis confirmed known roles of TFs involved in blood specification at key developmental stages but also identified signalling responsive TFs required for correct blood cell development, such as activator protein 1 (AP-1) and transcriptional enhanced associate domain (TEAD).
The hematopoietic cell fate is established by the interplay between signalling responsive and differentially expressed TFs
Becoming a blood cell requires the developmentally controlled expression of genes coding for TFs that are crucial for hematopoiesis, which interact with constitutively expressed and signalling responsive genes to control the transition from one GRN to another. In recent years, we have obtained significant insights into the mechanism of action of the most important hematopoietic TFs driving hematopoietic development and how they respond to signals.
The studies described above demonstrate that the final event in the generation of HSCs and multipotent progenitor cells is the EHT where blood cell fate is finally established. The EHT takes place in a defined cellular context. The site of the EHT in the midgestational mouse embryo is restricted to an endothelial cell layer at the ventral side of the dorsal aorta, demonstrating that the signalling environment plays an essential role in directing hematopoietic cell fate (5,45-48). These endothelial cells sit on the dorsal mesenchyme, which communicates through signalling to support and drive the commitment of endothelial cells towards the hematopoietic lineage between E8.5 and E10.5 (49-56). The TFs which direct this process can be broadly categorised into those that maintain an endothelial cell identity and those which drive the establishment of a hematopoietic cell fate. It is now clear from the study of the molecular events governing hematopoietic specification in vivo and in vitro that this process is highly complex and dynamic and is under tight transcriptional and signalling control (57). The next two paragraphs summarize the most important components.
The development and growth of endothelial cells and thus also the HE, requires the expression of the vascular endothelial growth factor receptor 2 (FLK-1) (28) which binds the vascular endothelial growth factor (VEGF) and induces signalling through the MAP kinase signalling pathway. TFs required for the development of these cells are SOX17, other members of the SOXF family (SOX7 and SOX18) and ETS TFs (ETV2) (58). At the same developmental stage, SOX17 is required for the repression of the hematopoietic genes Runx1 and Gata2 whose chromatin is already in a primed configuration (39,59). ETV2 deficiency results in a complete block of endothelial and hematopoietic cell type formation in knockout mouse models (60). An important signalling pathway regulating the EHT is NOTCH1 signalling. Loss-of-function studies indicate that it is required for definitive hematopoiesis, while gain-of-function studies found that intra-aortic clusters failed to form in the AGM indicating that NOTCH1 signalling needs to be switched off after the EHT (61). ETV2 and SOX17 establish and maintain paracrine NOTCH1 signalling between endothelial cells by upregulating the expression of NOTCH1 and its ligand DLL4 (62). Notch1 and Dll4 expression requires the TFs FOXC1 and FOXC2 in response to VEGF/PI3K-mediated signalling (63-65). FoxC2-null mouse embryos show impaired definitive hematopoiesis while zebrafish foxc1a/b morphants have reduced expression of hematopoietic genes (runx1, cmyb and rag1) in the HE (66).
NOTCH signalling is directly linked to transcriptional control and provides an example of a tight feedback mechanism in the control of cell fate decisions. Binding of the NOTCH1 receptor to its ligand expressed on a neighbouring endothelial cell exposes the NOTCH1 cleavage site S2 to the metalloprotease ADAM10, leading to cleavage. ADAM-10 deficient mice die at E9.5 with defects in their central nervous system, somites and cardiovascular system. They show reduced expression of the NOTCH target gene hes-5 and increased expression of the NOTCH ligand Dll-1 (67-70). After cleavage, the NOTCH1 intracellular domain (NICD) translocates into the nucleus, interacts with the TF RBPJ and turns it from a repressor into an activator. NICD/RBPJ then recruit mastermind like transcription coactivator (MAML) and histone acetyl transferases which up-regulate the expression of NOTCH target genes (71). NOTCH1 signalling to RBPJκ controls the activation of hematopoietic genes, for example, RBPJκ binds to and activates the promoter of Gata2 (72,73). In concordance with this result, RBPJκ mutant mouse embryos fail to form HSCs (72). However, NOTCH1 signalling also induces Hes genes encoding TFs that bind to and repress Gata2, thus preventing its overexpression once HES TFs reach a threshold level (72,74) thus maintaining endothelial identity.
Other important signalling pathways involved in the formation of endothelial cells and the HE are MAP Kinase signalling and HIPPO signalling, both of which regulate specific transcriptional programmes via signalling responsive TFs. MAP Kinase signalling terminates at the AP-1 family of TFs which consists of multiple JUN and FOS family proteins whereby FOS factors have to partner with JUN factors to be able to bind to DNA (75). The TF binding motif analysis performed by Goode et al., showed a specific enrichment of AP-1 binding motifs in the HE suggesting an important role for these factors in regulating the hemogenic cell fate (39). It was indeed shown that c-JUN knockout mice die at mid- to late-gestation due to impaired hepatogenesis and foetal liver erythropoiesis (76,77). JUND and FOS are required for hematopoiesis in Xenopus embryos (78) and JUNB is an important regulator of the EHT in the HE derived from human ESCs (79). It has been shown that VEGFA signals via AP-1 (80). Experiments from our lab used ChIP experiments to determine the position of JUN and FOS within the GRN (40). Moreover, induction of a dominant-negative (dn)FOS peptide at different stages of murine ESC derived hematopoietic differentiation which blocked all AP-1 binding activity showed that expression affected the development of endothelial cells (40) and modulates the balance between vascular smooth muscle and hemogenic cell fate.
HIPPO signalling which involves a large number of different signals controlling cell shape and cell communication, such as integrin signalling, sheer stress and many others [reviewed in (81)] has only recently been shown to be important for hematopoietic specification (39,82). The central components of this signalling pathway are the TEAD TFs and their co-activators YAP/TAZ. When Hippo signalling is active, YAP/TAZ are phosphorylated by a variety of different kinases, bind 14-3-3 proteins and are targeted for degradation. If HIPPO signalling is off, YAP/TAZ translocate into the nucleus, partner with TEAD and regulate gene expression. We have shown that the interaction between TEAD and YAP is essential for the EHT to occur (39). In addition, experiments from the North lab (82) showed that YAP responds to shear stress as it would be observed in the aorta and is required to up-regulate RUNX1 which initiates the hematopoietic program as explained in more detail below. Importantly, the dnFOS experiments (40) also revealed that TEAD and AP-1 cooperatively bind to specific cis-regulatory elements regulating endothelial and hematopoietic genes. Furthermore, they identified a sub-set of cis-regulatory elements where TEAD and AP-1 binding was interdependent to integrate MAP Kinase and HIPPO signalling at the genomic level (40). These experiments added to a growing body of literature that demonstrates that AP-1 is a common interaction partner driving a genomic response in the presence of multiple, but not individual signals (83).
GATA2 is a member of the GATA family of TFs (84) and is considered essential for the EHT for both primitive and definitive hematopoiesis. In the dorsal aorta, Gata2 deletion therefore results in a deficiency of intra-aortic clusters and HSCs in mouse embryos (85-88). Null mutations in Scl which encodes the TF SCL/TAL1 (89) cause a block of primitive erythropoiesis and homozygous mutant embryos die on E8.5 to E10.5. Furthermore, Scl (−/−) ESCs do not contribute to adult type hematopoiesis on chimeric analysis (90). However, the factor that truly drives the EHT is RUNX1 (91).
Runx1 is a master regulator of hematopoiesis and is essential for the EHT with Runx1 knockout resulting in a complete failure in HSC production from the HE (91). RUNX1 requires its cofactor CBFβ to complete the EHT as demonstrated by knockout of Cbfb which caused a similar phenotype to that seen in Runx1 knockout mouse embryos (92,93). The RUNX1 gene is transcribed by two promoters with differential developmental activity. The proximal promoter drives low-level expression leading to the binding of RUNX1 to down-stream targets in the HE and resulting in the up-regulation of cell adhesion- and migration-associated genes (94,95). Expression in the HE correlates with the binding of GATA2 and SCL/TAL1 which are already expressed (39,96). RUNX1 binds to its own cis-regulatory elements which strongly up-regulates Runx1 expression (97,98). Once RUNX1 levels pass a specific threshold, it reorganises the binding patterns of FLI-1 and SCL/TAL1 and the chromatin landscape of HE cells (97) and up-regulates other hematopoietic genes such as Spi1 (PU.1) and Cebpa (99). In parallel, it induces the expression of the repressive TFs GFI1 and GFI1B which cooperate with the co-repressor LSD1 to shut down endothelial TF gene expression (100). RUNX1 also directly binds to and represses Sox17 further shutting down endothelial-specific gene expression (97). In addition, RUNX1 expression changes the signalling environment by binding directly to the promoter of the Flk-1 (Kdr) gene and down-regulating its expression (101). Furthermore, RUNX1 represses Dll4 and Notch1 transcription (102), resulting in the loss of the VEGF-NOTCH signalling axis which maintains an endothelial cell signature and represses a hematopoietic signature (100,103). In parallel, and in cooperation with PU.1, RUNX1 up-regulates genes for hematopoietic cytokines such as GM-CSF, IL-1 and CSF1R thus driving hematopoietic differentiation forward (104,105). RUNX1 therefore creates a feed forward loop which drives blood cells development and growth. However, as described above, it is under strong repressive control by NOTCH1 and SOX17 in the HE, and the question then remained of how the balance of Runx1 activation and repression is regulated at the molecular level and which cis-regulatory elements are involved.
A genome-wide screen identifying developmentally regulated enhancer and promoter elements
To understand the mechanisms how dynamic GRNs control embryonic development processes we need to elucidate when and how cis-regulatory elements function at different developmental stages. For many decades, enhancer sequences were defined by transient or integrated reporter gene assays which show whether a specific sequence can stimulate the activity of a promoter independent of its position and distance (106). However, these assays do not inform on the dynamic behaviour of enhancers during development. In addition, enhancers were defined in a correlative way by the modification status of their flanking histones or by the fact that they are transcribed (107,108). The gold standard for experimentally studying the developmental activity of individual cis-regulatory elements are transgenic animal models which reveal the precise spatial and temporal activity of enhancers. However, these models are expensive and present challenges by integrating variable copy numbers of the transgene and displaying positional effects, requiring multiple transgenic lines to be produced to achieve a reproducible expression pattern (109,110). To study the role of individual enhancers within individual gene loci, they must be perturbed in their genomic environment using genome editing (107) but as with any candidate approach, such experiments only investigate one locus at the time.
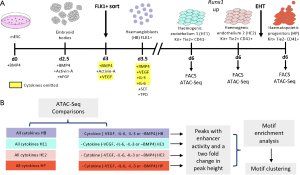
To rigorously study developmentally controlled cis-element activity in a high-throughput fashion and in a chromatin environment, we adapted the enhancer reporter system developed by Wilkinson et al. (109,111) to perform a genome-wide enhancer screen (112). We first differentiated mouse ESCs in the serum containing culture system (Figure 2A) and sorted cells by fluorescence-activated cell sorting (FACS), which were then taken for assay of transposase accessible chromatin sequencing (ATAC-seq) to identify open chromatin regions highlighting potentially active cis-elements. ATAC fragments were sequenced directly or cloned into a targeting vector to generate a fragment library which was then integrated into a defined target site in the HPRT locus carrying a minimal promoter to drive a reporter gene (Venus-YFP). Bulk mouse ESCs were differentiated into hematopoietic cells and cells from each stage of development were purified using cell sorting to measure reporter activity by flow cytometry (Figure 2B). Our enhancer screen returned several hundred thousand fragments which could stimulate the reporter construct; 22–31% of fragments mapped to distal elements and covered >70,000 ATAC sites differentially active across five cell stages. The rest of the fragments were promoter sequences which were defined as being ± 1.5 kb from an annotated transcription start site. Most of our positively scoring sequences overlapped with open chromatin sites found in purified endothelial cells and HE from mouse embryos at E9.5 and E13.5 (113,114). In addition, our screen picked up multiple cis-regulatory elements which had been previously identified within endogenous loci. Between 10% and 20% of all the distal sites and between 15% and 50% of all promoter sites displayed cell type specific activity in our screen and directed cell-type specific expression of their associated genes. In concordance with this finding, enhancer sequences were enriched in TF binding motifs specific for this particular differentiation stage with HE-specific enhancers displaying a TEAD/SOX/AP-1 motif signature which was replaced by a GATA/RUNX/ETS (PU.1) motif signature in HP-specific enhancers after the EHT. Multiple studies associated the presence of an active enhancer with specific types of histone modifications, such as H3K27 acetylation (115,116) or RNA-Polymerase II binding (117,118). Only half of the enhancers identified in our study were flanked by modified histones or were bound by RNA-Polymerase II, indicating that the absence of such features did not indicate an absence of enhancer activity.

Hematopoietic specification is controlled by a relay of signalling responsive enhancers
We next asked the question which of these enhancer elements were signalling responsive. To this end, we modified a serum-free differentiation system which employed the sequential addition of different cytokines to generate blood cell precursors (119) and omitted specific cytokines as shown in Figure 3 (112). We then used FACS to purify each cell type and measured chromatin accessibility in the presence and absence of these cytokines. These experiments showed that (I) thousands of cis-elements were cytokine responsive, (II) VEGF was the most important cytokine regulating the generation of blood cell precursor numbers and that (III) the presence of VEGF blocked the EHT. TF binding motif analysis of enhancer elements with and without cytokines demonstrated that in the presence of VEGF fewer cells containing open chromatin regions with motifs for hematopoietic TFs were formed. Cells were blocked at the endothelial stage as shown by an enriched endothelial TF motif signature. Interestingly, single cell RNA-Seq experiments showed that the actual differentiation pathway was not affected. Cells still underwent the correct succession of cell fate decisions from endothelial cells to HE to HP cells—what was different was the number of blood progenitors, suggesting that VEGF regulates a limiting factor operating at the EHT. This limiting factor turned out to be RUNX1 (112).
The Runx1 locus represents a signalling-responsive master switch driving the EHT
The single cell RNA-Seq experiments revealed that withdrawal of VEGF resulted in an up-regulation of Runx1 mRNA in HP cells and that in the presence of VEGF signalling the endothelial TF gene Sox17 was not efficiently down-regulated in these cells. In addition, the withdrawal of VEGF resulted in a significant decrease in the average mRNA expression values of Notch1. Moreover, the analysis of VEGF-responsive genes showed that the Dlk1 gene, which encodes a membrane bound repressor of NOTCH1 activity (120), was strongly up-regulated in the absence of VEGF, and that this gene was differentially expressed between HE clusters in the absence of VEGF, compared with HE clusters cultured with VEGF. These data support previous findings that VEGF establishes and maintains NOTCH1 signalling, and that VEGF signalling maintains the HE via ETV2 and SOX17 (60,121). VEGF therefore is a truly instructive cytokine which has a profound influence on the balance of expression of endothelial and hematopoietic TFs, resulting in an alteration of both the gene regulatory and signalling network.
To obtain insights into the molecular mechanisms by which this balance is regulated, we used our new data resource of functionally characterised cell-type specific enhancer elements to examine individual gene loci (112). First, we studied the Runx1 locus using our ATAC-seq data from cultures with and without VEGF. Our enhancer screen faithfully captured previously identified and validated Runx1 enhancer elements: the +23 kb enhancer (122), the +3.7 kb enhancer (113), a −371 kb enhancer (113), the +204 kb enhancer (123), the +110 kb enhancer (123) and others (123). Strikingly, open chromatin sites overlapping these enhancer elements only formed in the absence of VEGF signalling, whereas under both conditions promoters existed as open chromatin regions. Moreover, our ChIP and motif enrichment data which we accumulated over several years showed that these enhancer elements were bound by endothelial and hematopoietic TFs in the HE and hematopoietic cells, respectively (39-43). One of the strengths of the enhancer screening system is that it allows to study the cell stage-specific activity of individual wild type enhancer elements together with versions where specific binding motifs were mutated in the presence and absence of cytokines. These experiments demonstrated that VEGF operates via binding motifs for TEAD and AP-1 and is counteracted by RUNX1. TEAD factors repress Runx1 as, for example, at the Runx1 +23 kb enhancer its activity was strongly up-regulated when the TEAD site which binds TEAD4 as measured by ChIP was mutated, and down-regulated once an essential RUNX1 site (122) was eliminated. Once RUNX1 reaches a high level after VEGF withdrawal, it binds to its own enhancers, represses endothelial genes including Notch1 and activates hematopoietic genes.
The question now arises of the relevance of our in vitro data for mouse development. Recently Fadlullah et al. (33) used single cell (sc)-RNA-seq to capture the entire EHT process in mouse embryos focusing on the HE and dorsal aorta niche cells developing into intra-aortic hematopoietic clusters (IAHCs) which HP buds off from. Their data reveal a detailed HE differentiation continuum, which spanned the pre-HE and HE stages. When we mined the Fadlullah et al. data (33) we saw that, similar to what is seen in differentiating ESCs, Kdr (Flk-1) expression is highest in the endothelium and pre-EHT HE before expression is reduced dramatically in HE undergoing the EHT and IAHCs. Vegfa, which is expressed by the endothelium, also follows the same expression pattern. Conversely, Runx1 expression is low in endothelium and pre-EHT HE before being up-regulated in HE undergoing the EHT and in IAHCs. NOTCH genes such as Notch1, Dll1, Dll4, Jag1 and Jag2 were also down-regulated in HE undergoing the EHT and IAHCs compared to the endothelium. These findings are consistent with those from a study profiling the gene expression and chromatin accessibility profile of the mouse AGM around the emergence of the HSC between 9.5 to 11.5 days post coitus, revealing that Vegfa and Kdr expression was highest in non-HE and was reduced in HE and lowest in intra-aortic clusters and mature HSCs (113). A single-cell transcriptome map of human hematopoietic tissues generated from three 4.5–5 weeks old embryos (124) also showed that the expression of VEGF and its receptor KDR was also reduced in HSC populations compared to endothelium, while RUNX1 expression was highest in the HSC cell type.
These findings support our observations that Vegfa and Kdr expression is highest when Runx1 expression is low and this state switches in cells undergoing the EHT. Our studies of VEGF responsive enhancers now explain the molecular mechanisms governing how this occurs.
Outlook: using multi-omics data to understand developmental pathways
Our studies provide important mechanistic insights into the core regulatory and signalling network which regulates hematopoietic specification. They link extracellular signalling to the regulation of TF activity acting on specific cis-regulatory elements and shows that signalling has a profound impact on genomic events. Most importantly, they describe the dynamic activity of such elements within a developmental pathway adding an important dimension on our understanding of development. For example, we discovered multiple cis-regulatory elements which exist as open chromatin sites but lack enhancer activity in our assay [(39), Maytum, Edginton-White et al., in preparation]. Some of these sites become functional once the next developmental stage is reached, others do not, thus adding to the increased list of elements which appear to function solely in chromatin opening, thus facilitating the establishment of enhancers activated later in development [reviewed in (125)]. Another example for the usefulness of such data is in the ability to use motif analyses to investigate TF cooperation as exemplified in the cooperation of AP-1 and TEAD (40), with many other combinations suggested by the co-localization of their motifs. Our data is a valuable source for the study of how TF motifs and their arrangements regulate enhancer function and coupled with recently published methods such as (126) will provide insight into these combinations. Our enhancer lists, linked to promoter regions, may also be valuable in revealing how enhancers and promoters are paired and to which type of cis-element they belong which has been reported for different transcriptional programs (127-129). Finally, our data which link the activity of individual enhancers to the activity of its rightful gene enable the construction of mathematical models to predict additional enhancer elements (130,131) and the GRN behaviour which they direct in the presence or absence of signals (130,132,133). We believe that such studies are of the essence if we want to understand developmental processes and recapitulate them in vitro for regenerative medicine purposes.
Acknowledgments
Funding: This work was funded by grants from the Biotechnology and Biological Sciences Research Council (BBSRC: BB/R014809/1) and the Medical Research Council (MRC, MR/S021469/1) to CB and a BBSRC MiDTP studentship to AM.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at https://sci.amegroups.com/article/view/10.21037/sci-2023-011/coif). AM reports a MiDTP studentship from the Biotechnology and Biological Sciences Research Council, a salary from the Medical Research Council (MRC, MR/S021469/1) grant (to CB) and consultant fees from AMCA. BEW reports a salary from the Biotechnology and Biological Sciences Research Council (BBSRC: BB/R014809/1) grant (to CB) and consultant fees from AMCA. CB reports grants from the Biotechnology and Biological Sciences Research Council (BBSRC: BB/R014809/1), the Medical Research Council (MRC, MR/S021469/1) and consultant fees from AMCA.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Szabo Q, Bantignies F, Cavalli G. Principles of genome folding into topologically associating domains. Sci Adv 2019;5:eaaw1668.
- Davidson IF, Peters JM. Genome folding through loop extrusion by SMC complexes. Nat Rev Mol Cell Biol 2021;22:445-64. [Crossref] [PubMed]
- Abkowitz JL, Catlin SN, McCallie MT, et al. Evidence that the number of hematopoietic stem cells per animal is conserved in mammals. Blood 2002;100:2665-7. [Crossref] [PubMed]
- Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell 2008;132:631-44. [Crossref] [PubMed]
- de Bruijn MF, Speck NA, Peeters MC, et al. Definitive hematopoietic stem cells first develop within the major arterial regions of the mouse embryo. EMBO J 2000;19:2465-74. [Crossref] [PubMed]
- Keller G. Embryonic stem cell differentiation: emergence of a new era in biology and medicine. Genes Dev 2005;19:1129-55. [Crossref] [PubMed]
- Murry CE, Keller G. Differentiation of embryonic stem cells to clinically relevant populations: lessons from embryonic development. Cell 2008;132:661-80. [Crossref] [PubMed]
- Thomson JA, Itskovitz-Eldor J, Shapiro SS, et al. Embryonic stem cell lines derived from human blastocysts. Science 1998;282:1145-7. [Crossref] [PubMed]
- Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981;292:154-6. [Crossref] [PubMed]
- Martin GR. Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc Natl Acad Sci U S A 1981;78:7634-8. [Crossref] [PubMed]
- Aly RM. Current state of stem cell-based therapies: an overview. Stem Cell Investig 2020;7:8. [Crossref] [PubMed]
- Lee JY, Hong SH. Hematopoietic Stem Cells and Their Roles in Tissue Regeneration. Int J Stem Cells 2020;13:1-12. [Crossref] [PubMed]
- Bardot ES, Hadjantonakis AK. Mouse gastrulation: Coordination of tissue patterning, specification and diversification of cell fate. Mech Dev 2020;163:103617. [Crossref] [PubMed]
- Pijuan-Sala B, Griffiths JA, Guibentif C, et al. A single-cell molecular map of mouse gastrulation and early organogenesis. Nature 2019;566:490-5. [Crossref] [PubMed]
- Argelaguet R, Clark SJ, Mohammed H, et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 2019;576:487-91. [Crossref] [PubMed]
- Moore MA, Metcalf D. Ontogeny of the haemopoietic system: yolk sac origin of in vivo and in vitro colony forming cells in the developing mouse embryo. Br J Haematol 1970;18:279-96. [Crossref] [PubMed]
- Palis J, McGrath KE, Kingsley PD. Initiation of hematopoiesis and vasculogenesis in murine yolk sac explants. Blood 1995;86:156-63. [Crossref] [PubMed]
- Xu MJ, Matsuoka S, Yang FC, et al. Evidence for the presence of murine primitive megakaryocytopoiesis in the early yolk sac. Blood 2001;97:2016-22. [Crossref] [PubMed]
- Bertrand JY, Chi NC, Santoso B, et al. Haematopoietic stem cells derive directly from aortic endothelium during development. Nature 2010;464:108-11. [Crossref] [PubMed]
- Kingsley PD, Malik J, Fantauzzo KA, et al. Yolk sac-derived primitive erythroblasts enucleate during mammalian embryogenesis. Blood 2004;104:19-25. [Crossref] [PubMed]
- Palis J. Primitive and definitive erythropoiesis in mammals. Front Physiol 2014;5:3. [Crossref] [PubMed]
- Barone C, Orsenigo R, Meneveri R, et al. One Size Does Not Fit All: Heterogeneity in Developmental Hematopoiesis. Cells 2022;11:1061. [Crossref] [PubMed]
- Böiers C, Carrelha J, Lutteropp M, et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell 2013;13:535-48. [Crossref] [PubMed]
- Tavian M, Hallais MF, Péault B. Emergence of intraembryonic hematopoietic precursors in the pre-liver human embryo. Development 1999;126:793-803. [Crossref] [PubMed]
- Medvinsky A, Dzierzak E. Definitive hematopoiesis is autonomously initiated by the AGM region. Cell 1996;86:897-906. [Crossref] [PubMed]
- Boisset JC, van Cappellen W, Andrieu-Soler C, et al. In vivo imaging of haematopoietic cells emerging from the mouse aortic endothelium. Nature 2010;464:116-20. [Crossref] [PubMed]
- de Bruijn MF, Ma X, Robin C, et al. Hematopoietic stem cells localize to the endothelial cell layer in the midgestation mouse aorta. Immunity 2002;16:673-83. [Crossref] [PubMed]
- Jaffredo T, Gautier R, Eichmann A, et al. Intraaortic hemopoietic cells are derived from endothelial cells during ontogeny. Development 1998;125:4575-83. [Crossref] [PubMed]
- Kissa K, Herbomel P. Blood stem cells emerge from aortic endothelium by a novel type of cell transition. Nature 2010;464:112-5. [Crossref] [PubMed]
- Mizuochi C, Fraser ST, Biasch K, et al. Intra-aortic clusters undergo endothelial to hematopoietic phenotypic transition during early embryogenesis. PLoS One 2012;7:e35763. [Crossref] [PubMed]
- Tavian M, Coulombel L, Luton D, et al. Aorta-associated CD34+ hematopoietic cells in the early human embryo. Blood 1996;87:67-72. [Crossref] [PubMed]
- Zovein AC, Hofmann JJ, Lynch M, et al. Fate tracing reveals the endothelial origin of hematopoietic stem cells. Cell Stem Cell 2008;3:625-36. [Crossref] [PubMed]
- Fadlullah MZH, Neo WH, Lie-A-Ling M, et al. Murine AGM single-cell profiling identifies a continuum of hemogenic endothelium differentiation marked by ACE. Blood 2022;139:343-56. [Crossref] [PubMed]
- Rybtsov S, Batsivari A, Bilotkach K, et al. Tracing the origin of the HSC hierarchy reveals an SCF-dependent, IL-3-independent CD43(-) embryonic precursor. Stem Cell Reports 2014;3:489-501. [Crossref] [PubMed]
- Delassus S, Cumano A. Circulation of hematopoietic progenitors in the mouse embryo. Immunity 1996;4:97-106. [Crossref] [PubMed]
- Yzaguirre AD, Speck NA. Insights into blood cell formation from hemogenic endothelium in lesser-known anatomic sites. Dev Dyn 2016;245:1011-28. [Crossref] [PubMed]
- Cockerill PN. Structure and function of active chromatin and DNase I hypersensitive sites. FEBS J 2011;278:2182-210. [Crossref] [PubMed]
- Edginton-White B, Bonifer C. The transcriptional regulation of normal and malignant blood cell development. FEBS J 2022;289:1240-55. [Crossref] [PubMed]
- Goode DK, Obier N, Vijayabaskar MS, et al. Dynamic Gene Regulatory Networks Drive Hematopoietic Specification and Differentiation. Dev Cell 2016;36:572-87. [Crossref] [PubMed]
- Obier N, Cauchy P, Assi SA, et al. Cooperative binding of AP-1 and TEAD4 modulates the balance between vascular smooth muscle and hemogenic cell fate. Development 2016;143:4324-40. [Crossref] [PubMed]
- Gilmour J, Assi SA, Noailles L, et al. The Co-operation of RUNX1 with LDB1, CDK9 and BRD4 Drives Transcription Factor Complex Relocation During Haematopoietic Specification. Sci Rep 2018;8:10410. [Crossref] [PubMed]
- Gilmour J, O'Connor L, Middleton CP, et al. Robust hematopoietic specification requires the ubiquitous Sp1 and Sp3 transcription factors. Epigenetics Chromatin 2019;12:33. [Crossref] [PubMed]
- Kellaway SG, Keane P, Edginton-White B, et al. Different mutant RUNX1 oncoproteins program alternate haematopoietic differentiation trajectories. Life Sci Alliance 2021;4:e202000864. [Crossref] [PubMed]
- Neph S, Stergachis AB, Reynolds A, et al. Circuitry and dynamics of human transcription factor regulatory networks. Cell 2012;150:1274-86. [Crossref] [PubMed]
- Taoudi S, Medvinsky A. Functional identification of the hematopoietic stem cell niche in the ventral domain of the embryonic dorsal aorta. Proc Natl Acad Sci U S A 2007;104:9399-403. [Crossref] [PubMed]
- Ivanovs A, Rybtsov S, Welch L, et al. Highly potent human hematopoietic stem cells first emerge in the intraembryonic aorta-gonad-mesonephros region. J Exp Med 2011;208:2417-27. [Crossref] [PubMed]
- Medvinsky AL, Samoylina NL, Müller AM, et al. An early pre-liver intraembryonic source of CFU-S in the developing mouse. Nature 1993;364:64-7. [Crossref] [PubMed]
- Müller AM, Medvinsky A, Strouboulis J, et al. Development of hematopoietic stem cell activity in the mouse embryo. Immunity 1994;1:291-301. [Crossref] [PubMed]
- Richard C, Drevon C, Canto PY, et al. Endothelio-mesenchymal interaction controls runx1 expression and modulates the notch pathway to initiate aortic hematopoiesis. Dev Cell 2013;24:600-11. [Crossref] [PubMed]
- McGarvey AC, Rybtsov S, Souilhol C, et al. A molecular roadmap of the AGM region reveals BMPER as a novel regulator of HSC maturation. J Exp Med 2017;214:3731-51. [Crossref] [PubMed]
- Sá da Bandeira D, Kilpatrick AM, Marques M, et al. PDGFRβ+ cells play a dual role as hematopoietic precursors and niche cells during mouse ontogeny. Cell Rep 2022;40:111114. [Crossref] [PubMed]
- Ivanovs A, Rybtsov S, Anderson RA, et al. Identification of the niche and phenotype of the first human hematopoietic stem cells. Stem Cell Reports 2014;2:449-56. [Crossref] [PubMed]
- Chandrakanthan V, Rorimpandey P, Zanini F, et al. Mesoderm-derived PDGFRA(+) cells regulate the emergence of hematopoietic stem cells in the dorsal aorta. Nat Cell Biol 2022;24:1211-25. [Crossref] [PubMed]
- Mendes SC, Robin C, Dzierzak E. Mesenchymal progenitor cells localize within hematopoietic sites throughout ontogeny. Development 2005;132:1127-36. [Crossref] [PubMed]
- Oostendorp RA, Harvey KN, Kusadasi N, et al. Stromal cell lines from mouse aorta-gonads-mesonephros subregions are potent supporters of hematopoietic stem cell activity. Blood 2002;99:1183-9. [Crossref] [PubMed]
- Marshall CJ, Moore RL, Thorogood P, et al. Detailed characterization of the human aorta-gonad-mesonephros region reveals morphological polarity resembling a hematopoietic stromal layer. Dev Dyn 1999;215:139-47. [Crossref] [PubMed]
- Canu G, Ruhrberg C. First blood: the endothelial origins of hematopoietic progenitors. Angiogenesis 2021;24:199-211. [Crossref] [PubMed]
- Costa G, Mazan A, Gandillet A, et al. SOX7 regulates the expression of VE-cadherin in the haemogenic endothelium at the onset of haematopoietic development. Development 2012;139:1587-98. [Crossref] [PubMed]
- Lizama CO, Hawkins JS, Schmitt CE, et al. Repression of arterial genes in hemogenic endothelium is sufficient for haematopoietic fate acquisition. Nat Commun 2015;6:7739. [Crossref] [PubMed]
- Liu F, Li D, Yu YY, et al. Induction of hematopoietic and endothelial cell program orchestrated by ETS transcription factor ER71/ETV2. EMBO Rep 2015;16:654-69. [Crossref] [PubMed]
- Cortegano I, Melgar-Rojas P, Luna-Zurita L, et al. Notch1 regulates progenitor cell proliferation and differentiation during mouse yolk sac hematopoiesis. Cell Death Differ 2014;21:1081-94. [Crossref] [PubMed]
- Corada M, Orsenigo F, Morini MF, et al. Sox17 is indispensable for acquisition and maintenance of arterial identity. Nat Commun 2013;4:2609. [Crossref] [PubMed]
- Hayashi H, Kume T. Foxc transcription factors directly regulate Dll4 and Hey2 expression by interacting with the VEGF-Notch signaling pathways in endothelial cells. PLoS One 2008;3:e2401. [Crossref] [PubMed]
- Liu ZJ, Shirakawa T, Li Y, et al. Regulation of Notch1 and Dll4 by vascular endothelial growth factor in arterial endothelial cells: implications for modulating arteriogenesis and angiogenesis. Mol Cell Biol 2003;23:14-25. [Crossref] [PubMed]
- De Val S, Chi NC, Meadows SM, et al. Combinatorial regulation of endothelial gene expression by ets and forkhead transcription factors. Cell 2008;135:1053-64. [Crossref] [PubMed]
- Jang IH, Lu YF, Zhao L, et al. Notch1 acts via Foxc2 to promote definitive hematopoiesis via effects on hemogenic endothelium. Blood 2015;125:1418-26. [Crossref] [PubMed]
- Hartmann D, de Strooper B, Serneels L, et al. The disintegrin/metalloprotease ADAM 10 is essential for Notch signalling but not for alpha-secretase activity in fibroblasts. Hum Mol Genet 2002;11:2615-24. [Crossref] [PubMed]
- Weber S, Niessen MT, Prox J, et al. The disintegrin/metalloproteinase Adam10 is essential for epidermal integrity and Notch-mediated signaling. Development 2011;138:495-505. [Crossref] [PubMed]
- Gibb DR, El Shikh M, Kang DJ, et al. ADAM10 is essential for Notch2-dependent marginal zone B cell development and CD23 cleavage in vivo. J Exp Med 2010;207:623-35. [Crossref] [PubMed]
- van Tetering G, van Diest P, Verlaan I, et al. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem 2009;284:31018-27. [Crossref] [PubMed]
- Giaimo BD, Gagliani EK, Kovall RA, et al. Transcription Factor RBPJ as a Molecular Switch in Regulating the Notch Response. Adv Exp Med Biol 2021;1287:9-30. [Crossref] [PubMed]
- Robert-Moreno A, Espinosa L, de la Pompa JL, et al. RBPjkappa-dependent Notch function regulates Gata2 and is essential for the formation of intra-embryonic hematopoietic cells. Development 2005;132:1117-26. [Crossref] [PubMed]
- Butko E, Distel M, Pouget C, et al. Gata2b is a restricted early regulator of hemogenic endothelium in the zebrafish embryo. Development 2015;142:1050-61. [Crossref] [PubMed]
- Guiu J, Shimizu R, D'Altri T, et al. Hes repressors are essential regulators of hematopoietic stem cell development downstream of Notch signaling. J Exp Med 2013;210:71-84. [Crossref] [PubMed]
- Bejjani F, Evanno E, Zibara K, et al. The AP-1 transcriptional complex: Local switch or remote command? Biochim Biophys Acta Rev Cancer 2019;1872:11-23. [Crossref] [PubMed]
- Eferl R, Sibilia M, Hilberg F, et al. Functions of c-Jun in liver and heart development. J Cell Biol 1999;145:1049-61. [Crossref] [PubMed]
- Hilberg F, Aguzzi A, Howells N, et al. c-jun is essential for normal mouse development and hepatogenesis. Nature 1993;365:179-81. [Crossref] [PubMed]
- Lee SY, Yoon J, Lee MH, et al. The role of heterodimeric AP-1 protein comprised of JunD and c-Fos proteins in hematopoiesis. J Biol Chem 2012;287:31342-8. [Crossref] [PubMed]
- Chen X, Wang P, Qiu H, et al. Integrative epigenomic and transcriptomic analysis reveals the requirement of JUNB for hematopoietic fate induction. Nat Commun 2022;13:3131. [Crossref] [PubMed]
- Fan F, Malvestiti S, Vallet S, et al. JunB is a key regulator of multiple myeloma bone marrow angiogenesis. Leukemia 2021;35:3509-25. [Crossref] [PubMed]
- Zheng Y, Pan D. The Hippo Signaling Pathway in Development and Disease. Dev Cell 2019;50:264-82. [Crossref] [PubMed]
- Lundin V, Sugden WW, Theodore LN, et al. YAP Regulates Hematopoietic Stem Cell Formation in Response to the Biomechanical Forces of Blood Flow. Dev Cell 2020;52:446-460.e5. [Crossref] [PubMed]
- Macián F, López-Rodríguez C, Rao A. Partners in transcription: NFAT and AP-1. Oncogene 2001;20:2476-89. [Crossref] [PubMed]
- Katsumura KR, Bresnick EH. The GATA factor revolution in hematology. Blood 2017;129:2092-102. [Crossref] [PubMed]
- de Pater E, Kaimakis P, Vink CS, et al. Gata2 is required for HSC generation and survival. J Exp Med 2013;210:2843-50. [Crossref] [PubMed]
- Tsai FY, Keller G, Kuo FC, et al. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994;371:221-6. [Crossref] [PubMed]
- Gao X, Johnson KD, Chang YI, et al. Gata2 cis-element is required for hematopoietic stem cell generation in the mammalian embryo. J Exp Med 2013;210:2833-42. [Crossref] [PubMed]
- Ling KW, Ottersbach K, van Hamburg JP, et al. GATA-2 plays two functionally distinct roles during the ontogeny of hematopoietic stem cells. J Exp Med 2004;200:871-82. [Crossref] [PubMed]
- Shivdasani RA, Mayer EL, Orkin SH. Absence of blood formation in mice lacking the T-cell leukaemia oncoprotein tal-1/SCL. Nature 1995;373:432-4. [Crossref] [PubMed]
- Porcher C, Swat W, Rockwell K, et al. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell 1996;86:47-57. [Crossref] [PubMed]
- North T, Gu TL, Stacy T, et al. Cbfa2 is required for the formation of intra-aortic hematopoietic clusters. Development 1999;126:2563-75. [Crossref] [PubMed]
- Niki M, Okada H, Takano H, et al. Hematopoiesis in the fetal liver is impaired by targeted mutagenesis of a gene encoding a non-DNA binding subunit of the transcription factor, polyomavirus enhancer binding protein 2/core binding factor. Proc Natl Acad Sci U S A 1997;94:5697-702. [Crossref] [PubMed]
- Wang Q, Stacy T, Miller JD, et al. The CBFbeta subunit is essential for CBFalpha2 (AML1) function in vivo. Cell 1996;87:697-708. [Crossref] [PubMed]
- Sroczynska P, Lancrin C, Kouskoff V, et al. The differential activities of Runx1 promoters define milestones during embryonic hematopoiesis. Blood 2009;114:5279-89. [Crossref] [PubMed]
- Lie-A-Ling M, Marinopoulou E, Li Y, et al. RUNX1 positively regulates a cell adhesion and migration program in murine hemogenic endothelium prior to blood emergence. Blood 2014;124:e11-20. [Crossref] [PubMed]
- Nottingham WT, Jarratt A, Burgess M, et al. Runx1-mediated hematopoietic stem-cell emergence is controlled by a Gata/Ets/SCL-regulated enhancer. Blood 2007;110:4188-97. [Crossref] [PubMed]
- Lichtinger M, Ingram R, Hannah R, et al. RUNX1 reshapes the epigenetic landscape at the onset of haematopoiesis. EMBO J 2012;31:4318-33. [Crossref] [PubMed]
- Lie-A-Ling M, Marinopoulou E, Lilly AJ, et al. Regulation of RUNX1 dosage is crucial for efficient blood formation from hemogenic endothelium. Development 2018;145:dev149419. [Crossref] [PubMed]
- Zhang DE, Fujioka K, Hetherington CJ, et al. Identification of a region which directs the monocytic activity of the colony-stimulating factor 1 (macrophage colony-stimulating factor) receptor promoter and binds PEBP2/CBF (AML1). Mol Cell Biol 1994;14:8085-95. [PubMed]
- Lancrin C, Mazan M, Stefanska M, et al. GFI1 and GFI1B control the loss of endothelial identity of hemogenic endothelium during hematopoietic commitment. Blood 2012;120:314-22. [Crossref] [PubMed]
- Hirai H, Samokhvalov IM, Fujimoto T, et al. Involvement of Runx1 in the down-regulation of fetal liver kinase-1 expression during transition of endothelial cells to hematopoietic cells. Blood 2005;106:1948-55. [Crossref] [PubMed]
- Miller M, Chen A, Gobert V, et al. Control of RUNX-induced repression of Notch signaling by MLF and its partner DnaJ-1 during Drosophila hematopoiesis. PLoS Genet 2017;13:e1006932. [Crossref] [PubMed]
- Thambyrajah R, Mazan M, Patel R, et al. GFI1 proteins orchestrate the emergence of haematopoietic stem cells through recruitment of LSD1. Nat Cell Biol 2016;18:21-32. [Crossref] [PubMed]
- Zent CS, Mathieu C, Claxton DF, et al. The chimeric genes AML1/MDS1 and AML1/EAP inhibit AML1B activation at the CSF1R promoter, but only AML1/MDS1 has tumor-promoter properties. Proc Natl Acad Sci U S A 1996;93:1044-8. [Crossref] [PubMed]
- Zhang DE, Hohaus S, Voso MT, et al. Function of PU.1 (Spi-1), C/EBP, and AML1 in early myelopoiesis: regulation of multiple myeloid CSF receptor promoters. Curr Top Microbiol Immunol 1996;211:137-47. [Crossref] [PubMed]
- Banerji J, Rusconi S, Schaffner W. Expression of a beta-globin gene is enhanced by remote SV40 DNA sequences. Cell 1981;27:299-308. [Crossref] [PubMed]
- Field A, Adelman K. Evaluating Enhancer Function and Transcription. Annu Rev Biochem 2020;89:213-34. [Crossref] [PubMed]
- Heintzman ND, Stuart RK, Hon G, et al. Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 2007;39:311-8. [Crossref] [PubMed]
- Wilkinson AC, Goode DK, Cheng YH, et al. Single site-specific integration targeting coupled with embryonic stem cell differentiation provides a high-throughput alternative to in vivo enhancer analyses. Biol Open 2013;2:1229-38. [Crossref] [PubMed]
- Bonifer C. Developmental regulation of eukaryotic gene loci: which cis-regulatory information is required? Trends Genet 2000;16:310-5. [Crossref] [PubMed]
- Dickel DE, Zhu Y, Nord AS, et al. Function-based identification of mammalian enhancers using site-specific integration. Nat Methods 2014;11:566-71. [Crossref] [PubMed]
- Edginton-White B, Maytum A, Kellaway SG, et al. A genome-wide relay of signalling-responsive enhancers drives hematopoietic specification. Nat Commun 2023;14:267. [Crossref] [PubMed]
- Zhu Q, Gao P, Tober J, et al. Developmental trajectory of prehematopoietic stem cell formation from endothelium. Blood 2020;136:845-56. [Crossref] [PubMed]
- Howell ED, Yzaguirre AD, Gao P, et al. Efficient hemogenic endothelial cell specification by RUNX1 is dependent on baseline chromatin accessibility of RUNX1-regulated TGFβ target genes. Genes Dev 2021;35:1475-89. [Crossref] [PubMed]
- Creyghton MP, Cheng AW, Welstead GG, et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 2010;107:21931-6. [Crossref] [PubMed]
- Raisner R, Kharbanda S, Jin L, et al. Enhancer Activity Requires CBP/P300 Bromodomain-Dependent Histone H3K27 Acetylation. Cell Rep 2018;24:1722-9. [Crossref] [PubMed]
- Kim TK, Hemberg M, Gray JM, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature 2010;465:182-7. [Crossref] [PubMed]
- Lam MT, Li W, Rosenfeld MG, et al. Enhancer RNAs and regulated transcriptional programs. Trends Biochem Sci 2014;39:170-82. [Crossref] [PubMed]
- Pearson S, Sroczynska P, Lacaud G, et al. The stepwise specification of embryonic stem cells to hematopoietic fate is driven by sequential exposure to Bmp4, activin A, bFGF and VEGF. Development 2008;135:1525-35. [Crossref] [PubMed]
- Mirshekar-Syahkal B, Haak E, Kimber GM, et al. Dlk1 is a negative regulator of emerging hematopoietic stem and progenitor cells. Haematologica 2013;98:163-71. [Crossref] [PubMed]
- Casie Chetty S, Rost MS, Enriquez JR, et al. Vegf signaling promotes vascular endothelial differentiation by modulating etv2 expression. Dev Biol 2017;424:147-61. [Crossref] [PubMed]
- Bee T, Ashley EL, Bickley SR, et al. The mouse Runx1 +23 hematopoietic stem cell enhancer confers hematopoietic specificity to both Runx1 promoters. Blood 2009;113:5121-4. [Crossref] [PubMed]
- Owens DDG, Anselmi G, Oudelaar AM, et al. Dynamic Runx1 chromatin boundaries affect gene expression in hematopoietic development. Nat Commun 2022;13:773. [Crossref] [PubMed]
- Calvanese V, Capellera-Garcia S, Ma F, et al. Mapping human haematopoietic stem cells from haemogenic endothelium to birth. Nature 2022;604:534-40. [Crossref] [PubMed]
- Bonifer C, Cockerill PN. Chromatin priming of genes in development: Concepts, mechanisms and consequences. Exp Hematol 2017;49:1-8. [Crossref] [PubMed]
- Avsec Ž, Weilert M, Shrikumar A, et al. Base-resolution models of transcription-factor binding reveal soft motif syntax. Nat Genet 2021;53:354-66. [Crossref] [PubMed]
- Zabidi MA, Arnold CD, Schernhuber K, et al. Enhancer-core-promoter specificity separates developmental and housekeeping gene regulation. Nature 2015;518:556-9. [Crossref] [PubMed]
- Arnold CD, Zabidi MA, Pagani M, et al. Genome-wide assessment of sequence-intrinsic enhancer responsiveness at single-base-pair resolution. Nat Biotechnol 2017;35:136-44. [Crossref] [PubMed]
- Haberle V, Arnold CD, Pagani M, et al. Transcriptional cofactors display specificity for distinct types of core promoters. Nature 2019;570:122-6. [Crossref] [PubMed]
- de Almeida BP, Reiter F, Pagani M, et al. DeepSTARR predicts enhancer activity from DNA sequence and enables the de novo design of synthetic enhancers. Nat Genet 2022;54:613-24. [Crossref] [PubMed]
- Kleftogiannis D, Kalnis P, Bajic VB. Progress and challenges in bioinformatics approaches for enhancer identification. Brief Bioinform 2016;17:967-79. [Crossref] [PubMed]
- Beer MA, Shigaki D, Huangfu D. Enhancer Predictions and Genome-Wide Regulatory Circuits. Annu Rev Genomics Hum Genet 2020;21:37-54. [Crossref] [PubMed]
- Zehnder T, Benner P, Vingron M. Predicting enhancers in mammalian genomes using supervised hidden Markov models. BMC Bioinformatics 2019;20:157. [Crossref] [PubMed]
Cite this article as: Maytum A, Edginton-White B, Bonifer C. Identification and characterization of enhancer elements controlling cell type-specific and signalling dependent chromatin programming during hematopoietic development. Stem Cell Investig 2023;10:14.


