Ground state naïve pluripotent stem cells and CRISPR/Cas9 gene correction for β-thalassemia
The β-thalassemias are a group of hereditary diseases caused by more than 300 mutations of the adult β-globin gene, leading to low or absent production of adult hemoglobin (HbA) (1-3). Together with sickle cell anemia (SCA), thalassemia syndromes are among the most impactful diseases in developing countries, in which the lack of genetic counselling and prenatal diagnosis have contributed to the maintenance of a very high frequency in the population. The management of β-thalassemia patients is mostly based on blood transfusion, chelation therapy and, alternatively, on bone marrow transplantation (2). Recently, novel therapeutic options have been explored, such as gene therapy (3) and fetal hemoglobin (HbF) induction (4). Despite the fact that these approaches are promising, they are at present still under deep experimental development and limited to a low number of clinical trials (2-4). With respect to gene therapy for β-thalassemia significant progresses are expected, also considering fundamental insights into globin switching and new technology developments which might have a strong impact on novel gene-therapy approaches (3). A robust information is however available regarding the management of β-thalassemia, i.e., that patients exhibiting high levels of endogenous HbF might exhibit a milder clinical status, as in the case of hereditary persistence of fetal hemoglobin (HPFH) (4). In this context, one of the most exciting strategies recently proposed for hereditary diseases, including β-thalassemia, is genome editing using a variety of strongly validated approaches. Among these strategies, the clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system (5-7), in which a single guide RNA (sgRNA) directs the Cas9 nuclease for site-specific cleavage, is considered the most efficient.
Molecular therapies based on genome editing can be grouped in gene correction strategies or genetic disruption (8). The first step in both cases is based on the induction of a double strand break (DSB) by an engineered nuclease, which is endogenously repaired by homology-directed repair (HDR) or non-homologous end joining (NHEJ). Genetic correction strategies exploit the HDR pathway; the insertion of the corrected sequences (targeting specific gene mutations causing the molecular disease) is facilitated by the co-delivery of an extrachromosomal repair template in conjunction with the engineered nuclease, which improves the HDR frequency through the generation of DSB. By contrast, genetic disruption strategies are based on the NHEJ pathway following nuclease-induced DSB to produce local insertions/deletions (indels) (8).
As outlined in Figure 1, the combination of HSPC (hematological stem precursor cells) and iPSCs (induced pluripotent stem cells) production with gene correction strategies appears one of the most exciting and potentially therapeutic approaches for genetic diseases, including hematological pathologies, such as β-thalassemia and SCA (3). The creation of iPSCs from somatic cells with the use of reprogramming factors (originally Oct3/4, Sox2, c-Myc, and Klf4) (9) represented a key issue in our understanding of developmental biology and in the design of novel therapeutic approaches, also considering that their use avoids the ethical concerns associated with human embryonal stem cells (hESCs) and creates a patient-specific, histocompatible substrate for cell therapy. With respect of the importance of HbF production in the management of β-thalassemia, and since human iPSCs retain embryonic and fetal characteristics of gene expression even upon erythroid differentiation in vitro, one of the expected possibility was that patient-derived iPSCs for β-thalassemia or SCA (10-13) were able to maintain high levels of γ-globin expression (3). However, according to published in vivo findings after transplantation into immunodeficient mice, this very interesting possibility was not confirmed (14,15). Despite this apparent setback, iPSCs are a promising substrate for gene therapy, as they can be amplified in vitro indefinitely (where however they are still subject to the same mutation rates and potentially undesirable changes as any other cell type) and thus allow the clonal selection of guided events of therapeutic interest. Extensive use of iPSCs has been applied to the development of novel therapies for β-thalassemia and other hemoglobinopathies (3).
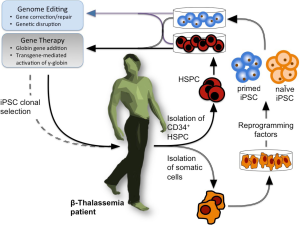
Induced pluripotent stem cells could be classified into two classes: primed iPSCs (12,13) and naïve iPSCs (16-20), both exhibiting therapeutic relevance. The naïve cells represent traits of mouse embryonic stem cells (mESC) derived from inner cell mass (ICM) or blastocyst from preimplantation embryo (18). Accordingly, naïve iPSCs exhibit molecular signatures of ground-state pluripotency with no tendency towards any lineage, retain an active X chromosome in female-derived iPSCs, are easier to be expanded and maintained, and differentiate along all the differentiation programs, due to the lack of lineage-commitment (18). On the contrary, primed iPSCs resemble cells derived from murine post-implantation epiblasts, exhibit low efficiency in cellular expansion and presence of activated lineage-commitment, thus preventing full differentiation potential (8).
In this context, a recent paper published by Yang et al. (21) on Stem Cell Translational Medicine clearly represents an important breakthrough. The relevance of the study of this group (from the Clinical and Translational Research Center of Shanghai, School of Life Sciences and Technology, Tongji University, China) is in the fact that they used the established 5i/L/FA system (18) to reprogram for the first time fibroblasts of a patient with β-thalassemia into transgene-free naïve iPSCs with molecular signatures of ground-state pluripotency. Using the CRISPR/Cas9 genome editing system, it was shown that these naïve iPSCs exhibited significantly improved gene-correction efficiencies compared with the corresponding primed iPSCs.
Gene editing on iPSCs: general considerations
The relevant parameters to be considered in iPSC gene editing are (I) the source of adult cells for iPSCs production; (II) the method for iPSCs production; (III) the genome editing system; (IV) the target genomic sequence(s) (8). These parameters impact with some features of the protocol, i.e., invasiveness of the sampling from β-thalassemia patients (point “I”); efficiency of iPSC production and growth ability (point “II”); off-target effects, genotoxicity and gene correction efficiency (point “III”); type of strategy for recovery the biological activity of β-thalassemia erythroid cells by correcting the primary mutation, modify regulatory region, or both (point “IV”).
Production of naïve iPSCs and advantages of their use
In the procedure presented by Yang et al. (21) fibroblasts were used for naïve iPSC production. For transfection, episomal vectors were employed encoding Oct3, Sox2, Klf4, Myc and NANOG, i.e., factors that play a pivotal role in iPSCs generation, as originally reported by the pioneering work by Takahashi & Yamanaka (9). The episomal vectors allow a transgene-free strategy for the expression of iPSC inducing factors, minimizing the risk of genotoxicity associated with integration in the genome of target cells. After transfection, the cells were cultured in slightly modified conventional human embryonic stem cell medium (hESM) for 6 days. Then, the medium was replaced with human naïve medium (5i/L/FA medium) and cultured for 14–20 days. During this culture period dome-shaped colonies similar to mouse ESCs were clearly visible and were selected and expanded by single cell passaging. As far the source of adult stem cells, Yang et al. used adult fibroblast and urinary cells (21). Importantly, as published by several research teams, also other cell types can be reprogrammed into a ground state, such as adipose stem cells, peripheral blood cells and hepatocytes (16-19). Notably, the use of urinary cells can be considered as a non-invasive technology (21).
The advantages of using naïve iPSCs are under debate and can be summarized as follows when comparison is made to primed iPSCs: high proliferation ability, efficient single-cell cloning and recovery. These features facilitate mutant gene targeting and drug screening, allowing application of iPSCs in disease modeling and regenerative medicine. Moreover naïve iPSCs can efficiently produce cross-species chimeras, allowing the development of very appealing experimental model systems (21).
Gene editing: the choice of the strategy
The relative benefits of alternative gene editing systems is at present under debate and should be carefully monitored with respect to several parameters, of which the most important are efficiency and off-target effects. The genome editing system employed by Yang et al. (21) for genetic correction of the the naïve iPSCs was the CRISPR/Cas9 strategy; however, other approaches can be considered, such as those based on zinc finger nucleases (ZFNs) (22) and TAL effector nucleases (TALENs) (23). Moreover, other Cas9-like systems have been described, including the CRISPR/Cpf1 nuclease platform, dimeric RNA-guided FokI nucleases, and use of Cas9’s derived from a variety of prokaryotic species (8).
Gene editing: the choice of the genomic target
In the procedure developed by Yang et al. (21) the target genomic sequence was the β-41/42 mutation of the β-globin gene (HBB). This is characterized by a TCTT deletion between the 41st and 42nd amino acids of the HBB gene, causing a frame-shift leading to a β0-thalassemia phenotype. It is expected that this approach will be useful for any other β-thalassemia mutations, with the exception of large gene deletions in which globin gene addition (instead of globin gene correction) is required.
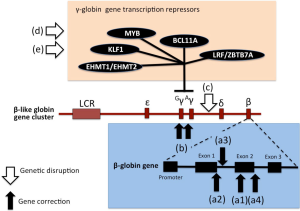
While the CRISPR/CAS gene correction was directed to the β-thalassemia mutation, the described strategy might also consider targeting of other genomic regions involved in key processes, such as production of fetal hemoglobin and generation of a HPFH-like phenotype. It should be underlined that genome editing-based therapies include not only gene correction but also disruption of target gene regions. In the case of β-thalassemia this gene disruption targeting might include not only discrete regions of the β-globin gene cluster, but also direct or indirect transcriptional repressors of human γ-globin genes, such as BCL11A, MYB, HLF-1, EHMT1/EHMT2 and LRF/ZBTB7A (see Figure 2). The description of these alternative targets in customizing gene editing has been recently reviewed by Canver and Orkin (8).
Combination therapy based on gene addition, gene editing and HbF inducers
As already pointed out, induction of endogenous HbF is one on the most widely applied therapeutic strategies for β-thalassemia and SCA (4). While most of the recent studies in the field still focus on small-molecular-weight HbF inducers, more recently the innovative strategy of combining vector-derived β-globin with the induction of endogenous HbF has been investigated. The combined treatment induces an increase of both HbA (by gene addition) and HbF (by chemical HbF induction), with important therapeutic implications, given that gene augmentation in β-thalassemia major has been unable to reach physiological levels of β-like globin to date and might thus only lead to partial phenotypic correction (24). Since increased production of HbF is beneficial in β-thalassemia, the one-off application of gene therapy implemented with chronic application of HbF inducers appears to be a pertinent strategy to achieve clinical benefits not achievable with either strategy alone. When gene therapy was combined to HbF induction, the results obtained demonstrate that this combination strategy achieves high levels of functional hemoglobin in β-thalassemic cells and a concomitant sharp decrease of excess α-globin, with significant scope for further improvements for what is as yet a nascent field of research. This combined approach, at least in theory, also applies considering gene editing and HbF induction.
CRISPR/Cas9 and iPSCs technologies: from the laboratory to the clinic
Despite the promising developments of CRISPR-based methodologies, many challenges have to be overcome before the system can be applied therapeutically to human patients, enabling delivery technology being one of the key issues. With respect to consideration on clinical trial, ZFNs are the editing technologies considered in protocols fighting several pathologies. However, recent developments concern also the CRISPR/Cas9 approach, considered as the key genetic editing system in four clinical trials focusing on the use of PD-1 knockout engineered T cells for metastatic non-small cell lung cancer (clinicaltrials.gov NCT02793856) and renal cell carcinoma (clinicaltrials.gov NCT02867332), hormone refractory prostate cancer (clinicaltrials.gov NCT02867345) and invasive bladder cancer Stage IV (clinicaltrials.gov NCT02863913). In these clinical trials CRISPR/Cas9 is proposed to neutralize the PD-1 gene, which expresses a protein on T-cell surfaces that many cancers can turn off, thereby blocking T-cell antitumor attacks. Once expanded in the laboratory, the engineered cells will be returned to the patient, circulate and hopefully home in on the cancer site.
On the other hand, the use of iPSCs in clinic is the object of a limited number of trials (more than 15 in clinicaltrials.gov), such as NCT00953693 (Patient Specific Induced Pluripotency Stem Cells), NCT02469207 (Regenerative Cellular Therapies, Physiology, Pathology and Developmental Biology), NCT02162953 (Stem Cell Models of Best Disease and Other Retinal Degenerative Diseases) and NCT02056613 (Blood Collection From Healthy Volunteers and Patients for the Production of Clinical Grade Induced Pluripotent Stem Cell Products).
The CRISPR/Cas9 patent war
When it was clear that the CRISPR technology was easy to be adapted to mammalian cells for developing novel and very impactful therapeutic options, the interest in patenting both approach and biomedical applications increased dramatically. The interest on CRISPR/Cas9 technology was the background for a spectacular patent war (25). Up to now the U.S. Patent and Trademark Office has granted more than 25 patents with claims concerning CRISPR and/or Cas9; among the more active institution and affiliated researchers is the Broad Institute, MIT, USA. Examples of granted patents are USP8871445 (CRISPR-Cas component systems, methods and compositions for sequence manipulation), USP9102936 (Method of adaptor-dimer subtraction using a CRISPR-CAS6 protein), USP8771945 (CRISPR-Cas systems and methods for altering expression of gene products) and USP8932814 (CRISPR-Cas nickase systems, methods and compositions for sequence manipulation in eukaryotes). In addition to these granted patents, a huge number of patent applications have been filed. This patent war competition between biotech companies is aimed to get the first CRISPR system to the therapeutic market and, as expected, involves also academic institutions and research groups involved in the novel developments of specific applications of the CRISPR technology (25). At present the dispute concerning the CRISPR/Cas9 patenting continues and might be responsible for a significant delay in clinical applications.
Acknowledgements
The authors apologize for the several original studies and reviews that were not cited, due to space limitation.
In memory: This work is dedicated to our colleague and friend Dr. Chiara Gemmo.
Funding: R Gambari is supported by the UE FP7 THALAMOSS Project (THALAssaemia MOdular Stratification System for personalized therapy of beta-thalassemia; grant no. 306201-FP7-Health-2012-INNOVATION-1) and by Fondazione Cariparo (Cassa di Risparmio di Padova e Rovigo).
Footnote
Provenance: This is a Guest Editorial commissioned by Editor-in-Chief Zhizhuang Joe Zhao (Pathology Graduate Program, University of Oklahoma Health Sciences Center, Oklahoma City, USA).
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Weatherall DJ. Phenotype-genotype relationships in monogenic disease: lessons from the thalassaemias. Nat Rev Genet 2001;2:245-55. [Crossref] [PubMed]
- Finotti A, Borgatti M, Bianchi N, et al. Orphan Drugs and Potential Novel Approaches for Therapies of β-Thalassemia: Current Status and Future Expectations. Expert Opin Orphan Drugs 2016;4:299-315. [Crossref]
- Finotti A, Breda L, Lederer CW, et al. Recent trends in the gene therapy of β-thalassemia. J Blood Med 2015;6:69-85. [PubMed]
- Finotti A, Gambari R. Recent trends for novel options in experimental biological therapy of β-thalassemia. Expert Opin Biol Ther 2014;14:1443-54. [Crossref] [PubMed]
- Traxler EA, Yao Y, Wang YD, et al. A genome-editing strategy to treat β-hemoglobinopathies that recapitulates a mutation associated with a benign genetic condition. Nat Med 2016;22:987-90. [Crossref] [PubMed]
- Xu P, Tong Y, Liu XZ, et al. Both TALENs and CRISPR/Cas9 directly target the HBB IVS2-654 (C > T) mutation in β-thalassemia-derived iPSCs. Sci Rep 2015;5:12065. [Crossref] [PubMed]
- Niu X, He W, Song B, et al. Combining Single Strand Oligodeoxynucleotides and CRISPR/Cas9 to Correct Gene Mutations in β-Thalassemia-induced Pluripotent Stem Cells. J Biol Chem 2016;291:16576-85. [Crossref] [PubMed]
- Canver MC, Orkin SH. Customizing the genome as therapy for the β-hemoglobinopathies. Blood 2016;127:2536-45. [Crossref] [PubMed]
- Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006;126:663-76. [Crossref] [PubMed]
- Ye L, Chang JC, Lin C, et al. Induced pluripotent stem cells offer new approach to therapy in thalassemia and sickle cell anemia and option in prenatal diagnosis in genetic diseases. Proc Natl Acad Sci U S A 2009;106:9826-30. [Crossref] [PubMed]
- Breda L, Gambari R, Rivella S. Gene therapy in thalassemia and hemoglobinopathies. Mediterr J Hematol Infect Dis 2009;1:e2009008. [PubMed]
- Wang Y, Zheng CG, Jiang Y, et al. Genetic correction of β-thalassemia patient-specific iPS cells and its use in improving hemoglobin production in irradiated SCID mice. Cell Res 2012;22:637-48. [Crossref] [PubMed]
- Fan Y, Luo Y, Chen X, et al. Generation of human β-thalassemia induced pluripotent stem cells from amniotic fluid cells using a single excisable lentiviral stem cell cassette. J Reprod Dev 2012;58:404-9. [Crossref] [PubMed]
- Ochi K, Takayama N, Hirose S, et al. Multicolor staining of globin subtypes reveals impaired globin switching during erythropoiesis in human pluripotent stem cells. Stem Cells Transl Med 2014;3:792-800. [Crossref] [PubMed]
- Dias J, Gumenyuk M, Kang H, et al. Generation of red blood cells from human induced pluripotent stem cells. Stem Cells Dev 2011;20:1639-47. [Crossref] [PubMed]
- Hanna JH. The STATs on naive iPSC reprogramming. Cell Stem Cell 2010;7:274-6. [Crossref] [PubMed]
- Nishishita N, Shikamura M, Takenaka C, et al. Generation of virus-free induced pluripotent stem cell clones on a synthetic matrix via a single cell subcloning in the naïve state. PLoS One 2012;7:e38389. [Crossref] [PubMed]
- Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014;15:471-87. [Crossref] [PubMed]
- Sanal MG. A highly efficient method for generation of therapeutic quality human pluripotent stem cells by using naive induced pluripotent stem cells nucleus for nuclear transfer. SAGE Open Med 2014;2:2050312114550375. [Crossref] [PubMed]
- Ou Z, Niu X, He W, et al. The Combination of CRISPR/Cas9 and iPSC Technologies in the Gene Therapy of Human β-thalassemia in Mice. Sci Rep 2016;6:32463. [Crossref] [PubMed]
- Yang Y, Zhang X, Yi L, et al. Naïve Induced Pluripotent Stem Cells Generated From β-Thalassemia Fibroblasts Allow Efficient Gene Correction With CRISPR/Cas9. Stem Cells Transl Med 2016;5:8-19. [Crossref] [PubMed]
- Ma N, Shan Y, Liao B, et al. Factor-induced Reprogramming and Zinc Finger Nuclease-aided Gene Targeting Cause Different Genome Instability in β-Thalassemia Induced Pluripotent Stem Cells (iPSCs). J Biol Chem 2015;290:12079-89. [Crossref] [PubMed]
- Ma N, Liao B, Zhang H, et al. Transcription activator-like effector nuclease (TALEN)-mediated gene correction in integration-free β-thalassemia induced pluripotent stem cells. J Biol Chem 2013;288:34671-9. [Crossref] [PubMed]
- Zuccato C, Breda L, Salvatori F, et al. A combined approach for β-thalassemia based on gene therapy-mediated adult hemoglobin (HbA) production and fetal hemoglobin (HbF) induction. Ann Hematol 2012;91:1201-13. [Crossref] [PubMed]
- Sherkow JS. Law, history and lessons in the CRISPR patent conflict. Nat Biotechnol 2015;33:256-7. [Crossref] [PubMed]
Cite this article as: Finotti A, Borgatti M, Gambari R. Ground state naïve pluripotent stem cells and CRISPR/Cas9 gene correction for β-thalassemia. Stem Cell Investig 2016;3:66.


