Memories of an X-chromosome
Heterogametic sex determination systems, where one sex is homogametic –XX or ZZ– and the other is heterogametic –XY or ZW–, entail a transcriptional dosage problem: in the absence of a compensatory mechanism, the homogametic sex expresses a double dose of those genes allocated on the duplicated sex chromosome. Eutherian mammals have evolved an epigenetic-based compensatory mechanism, termed X-chromosome inactivation (XCI) (1) that ensures that most X-linked genes are equally expressed in male (XY) and female (XX) individuals. In these species, XCI occurs randomly in the paternal or maternally inherited X-chromosome in such a way that females can be considered as mosaic individuals, expressing the paternally inherited X-chromosome in half of their cells and the maternally inherited one in the other half. The random nature of XCI confers an evolutionary advantage to females, as deleterious or detrimental mutations on one of the X-chromosomes, which can be somehow compensated by the cells expressing the “healthy” allele. This situation has a clear implication on some X-linked genetic disorders, which are lethal on males, being only manifested in females, and opens a way for the development of therapeutic approaches where those cells expressing the deleterious allele could be substituted or converted to cells expressing the “healthy” allele. Furthermore, the inactivation of a whole chromosome by XCI-based mechanisms has been successfully employed to correct transcriptional dosage in trisomy 21, responsible for Down’s syndrome (2). On the other hand, failures on XCI upon differentiation of XX human pluripotent stem cells (hPSCs) may hamper its therapeutic use due to aberrant expression of X-linked genes. In this perspective, the development of therapeutic applications requires substantial basic research on the dynamic epigenetic mechanisms leading to the establishment and reversion of XCI, both in embryos and stem cells. On January issue of Cell Stem Cell (3), Sahakyan et al. report a method to erase XCI abnormalities in primed hPSCs and have uncovered a yet uncharacterized epigenetic memory of XCI in hPSCs that should be considered in therapies aiming to switch transcription from the X-chromosome expressing a deleterious allele to the “healthy” one (Figure 1).
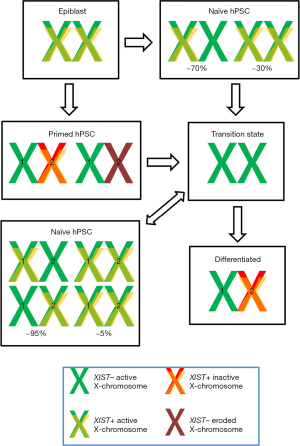
XCI entails a series of epigenetic mechanisms designed to inactivate one of the two X-chromosomes. The process is initiated by the expression of the long noncoding RNA XIST, which coats the cis X-chromosome. Following coating, certain proteins are recruited, including the histone variant macroH2A, ASHL2, SAFA and polycomb group proteins from the repressive complex 2. These recruited proteins alter the epigenetic landscape of the X-chromosome by modifying its epigenetic marks. The most characteristic epigenetic modification is the enrichment on H3K27me3, a repressive modification of histone 3 that excludes the binding of RNA-pol II, which is followed by DNA methylation, a more stable repressive mark. All these dynamic epigenetic changes occur during early embryogenesis, being concomitant with early cell differentiation processes; thereby XCI and reactivation are inherently bound to cell differentiation and reprogramming processes, respectively. In particular, primed pluripotent cells exhibit one inactive X-chromosome (XaXi), but reactivate the Xi when they are converted to the naïve type (XaXa). These changes mimic the XCI status of the cells from which they are derived in mice: early epiblast (XaXa) for naïve cells and late epiblast (XaXi) for primed cells. In contrast, hPSCs were initially found to be highly heterogeneous for this parameter (4), suggesting that there may be species-specific differences in XCI in stem cells, which may be rooted to differences in early development.
The laboratory mouse was the first model where XCI dynamics during early development was studied in detail. In this species, both X-chromosomes are active after embryonic genome activation, but later the paternal X-chromosome (Xp) is inactivated on early embryo cleavage stages. Then, during blastocyst formation, Xp remain inactive in trophectoderm (TE) cells, whereas epiblast cells experience a reactivation of the Xp, containing two Xa that remain active until gastrulation, where random inactivation occurs (5). The mouse model was assumed to be representative of other mammals, including humans, but later studies suggested that it is more an exception than a rule (6). Transcriptional studies in bovine, a species with preimplantation development timing similar to human, observed that XCI was far from being accomplished at the blastocysts stage (7), occurring later, during gastrulation (8). Similar findings were found in human blastocysts, where most blastomeres, including those of the TE, show two Xa that, in contrast to the mouse early epiblast, do express XIST from both Xa (9). The reason for XIST expression in Xa is unclear, it could be reasoned that it poises either of the X-chromosomes for later inactivation or that it serves to dampen the expression of both X-chromosomes. One article published last year suggests that XIST expression in human blastocysts may play a role in dampening X-chromosome expression, i.e., reducing the expression of both X chromosomes rather than inactivating one of them. The analysis of the transcriptome during human preimplantation development by RNAseq in single blastomeres revealed that both X-chromosome remain active throughout the whole period, as biallelic expression was maintained (10). However, they observed a progressive dampening of both X-chromosomes as (I) the sex-specific differences on the transcription of X-linked genes are gradually reduced; and (II) there is a female-specific progressive downregulation of X-linked genes. This gradual X chromosome dampening is accompanied by a steady upregulation of XIST (10). Together, different findings suggest that XCI in human embryos do not occur as previously thought, so the concept of “proper” XCI in hPSCs should be revisited.
The presence of two Xa in hPSCs has been suggested to be a hallmark of full pluripotency, as it resembles the situation found in the early epiblast (11). In agreement, early studies on conversion of primed hPSCs to naïve hPSCs suggested a reactivation of the Xi based on the absence of XIST cloud (12). However, as mentioned above, these cells did not mirror the situation of the human blastocyst (XaXa with two XIST clouds), uncovered one year later (9). Sahakyan et al. (3) achieve XCI status closer to that found in human blastocysts using 5iLAF conditions (13): most naïve cells derived from primed hPSCs lines or straight from blastocysts were XaXa with one cloud of XIST and some were XaXIST+XaXIST+ as the ICM blastomeres from which they originally derived (3). The incomplete recapitulation of XCI status in hPSCs, also manifested by an accumulation of H3K27me3 in naïve cells not observed in blastocysts, suggests that further improvement can be achieved. Nevertheless, the partial XCI reprogramming accomplished under 5iLAF conditions was able to erase Xi abnormalities of primed hPSCs. In particular, the authors were able to induce XCI following differentiation from the naïve state in some hPSCs lines that failed to achieve XCI when the differentiation started from primed cells (14).
Besides this achievement, the article reveals other interesting and unexpected observations. First, the authors suggest that Xi reactivation could rely on an X-specific mechanism, as demethylation in the Xi occurred earlier than in other genomic regions. Second, they report that both X-chromosome reactivation in the primed to naïve conversion and XCI in the differentiation of naïve cells transition through an XIST negative XaXa state. The loss of XIST during early differentiation suggests that XIST expression in naïve XaXa cells do not to poise them for later inactivation, therefore, XIST function seems to be limited to reduce the expression of both Xa, as suggested in human blastocysts based on the opposite transcriptional dynamics of XIST and other X-linked genes (10). In agreement, XaXa XIST positive cells exhibit similar extent of X-linked gene expression than XaXi primed cells. However, the role of XIST in X-chromosome dampening is also controversial, as most naïve cells showed monoallelic XIST expression, so dampening should only affect one X-chromosome, a question that remains unresolved in the manuscript. The XIST negative XaXa transitional state is not observed when naïve cells are derived straight from human blastocysts, but it can be observed in the derivation of primed cells from human blastocysts (14), so a similar transition may occur in the developing human embryo undergoing XCI.
Last, the most striking phenomenon is that XCI was not random in the reprogramed XaXa cells: following differentiation almost all naïve XaXa cells derived from XaXi cells inactivated the X-chromosome that was originally inactive. The epigenetic mark responsible for retaining memory on the reactivated Xi remains elusive, as the transcriptional status of both Xa was similar, XACT was biallelically expressed, and XIST was expressed by either of the Xa and transitionally lost during Xi reactivation and Xa inactivation. However, although some of the naïve XaXa cells do express XIST from the initially active Xa, a majority of the cells (57%) expressed only the same XIST allele than the primed XaXi cells, which may suggest that although XIST is not the stable mark responsible for XCI memory, the XCI memory somehow favours XIST expression. A priori epigenetic candidates for XCI memory are residual DNA methylation at specific sequences or histone modifications. The epigenetic memory of the reactivated Xa must be taken into consideration in therapeutic approaches: were hPSCs intended to differentiate into cells expressing the “healthy” allele, clones derived from cells expressing the “healthy” allele must be selected, a relatively easy task. However, the retention of this memory suggests that XCI status was not fully reversed and future approaches aiming to switch the expression to the “healthy” X-chromosome in situ would need to deal with the erasure of the unknown epigenetic memory for the prior Xi. The discovery of the epigenetic mechanism behind this memory may be also useful in the study of the epigenetic transgenerational inheritance, a phenomenon that is yet to be molecularly explained; the X-chromosome is still a good mine to dig up epigenetic answers.
Acknowledgements
Funding: Funding was provided by the projects RYC-2012-10193 and AGL2014-58739-R from the Spanish Ministry of Economy and Competitiveness. ILT is supported by an FPI contract.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961;190:372-3. [Crossref] [PubMed]
- Jiang J, Jing Y, Cost GJ, et al. Translating dosage compensation to trisomy 21. Nature 2013;500:296-300. [Crossref] [PubMed]
- Sahakyan A, Kim R, Chronis C, et al. Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell 2017;20:87-101. [Crossref] [PubMed]
- Silva SS, Rowntree RK, Mekhoubad S, et al. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A 2008;105:4820-5. [Crossref] [PubMed]
- Kay GF, Barton SC, Surani MA, et al. Imprinting and X chromosome counting mechanisms determine Xist expression in early mouse development. Cell 1994;77:639-50. [Crossref] [PubMed]
- Bermejo-Alvarez P, Ramos-Ibeas P, Gutierrez-Adan A. Solving the "X" in embryos and stem cells. Stem Cells Dev 2012;21:1215-24. [Crossref] [PubMed]
- Bermejo-Alvarez P, Rizos D, Rath D, et al. Sex determines the expression level of one third of the actively expressed genes in bovine blastocysts. Proc Natl Acad Sci U S A 2010;107:3394-9. [Crossref] [PubMed]
- Bermejo-Alvarez P, Rizos D, Lonergan P, et al. Transcriptional sexual dimorphism in elongating bovine embryos: implications for XCI and sex determination genes. Reproduction 2011;141:801-8. [Crossref] [PubMed]
- Okamoto I, Patrat C, Thepot D, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 2011;472:370-4. [Crossref] [PubMed]
- Petropoulos S, Edsgard D, Reinius B, et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016;165:1012-26. [Crossref] [PubMed]
- Navarro P, Avner P. When X-inactivation meets pluripotency: an intimate rendezvous. FEBS Letters 2009;583:1721-7. [Crossref] [PubMed]
- Hanna J, Cheng AW, Saha K, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A 2010;107:9222-7. [Crossref] [PubMed]
- Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014;15:471-87. [Crossref] [PubMed]
- Patel S, Bonora G, Sahakyan A, et al. Human Embryonic Stem Cells Do Not Change Their X Inactivation Status during Differentiation. Cell Rep 2017;18:54-67. [Crossref] [PubMed]
Cite this article as: Lamas-Toranzo I, Pericuesta E, Bermejo-Álvarez P. Memories of an X-chromosome. Stem Cell Investig 2017;4:27.


