X-chromosome activity in naive human pluripotent stem cells—are we there yet?
Introduction
A major goal for stem cell research and its clinical applications is to derive high quality human pluripotent stem cell (hPSC) lines that recapitulate in culture the properties of epiblast cells from human blastocyst embryos. Recent developments in derivation and culture conditions have allowed establishing so-called naive pluripotent hPSCs, which mimic closely the in vivo state on multiple levels like gene expression and differentiation potential. An epigenetic hallmark associated with naive pluripotency in female mouse cells is the reactivation of the X-chromosome and it was believed that this would be also the case for human naive cells. However, new evidence accumulates showing that the situation is not quite as simple. In this perspective, we describe the latest developments on this question and focus in particular on two recent studies by Sahakyan et al. and Vallot et al. (1,2), as they describe for the first time in detail the X-chromosome state of naive hPSCs. They provide important ground-work for studying human X-chromosome dynamics in vitro and for using the epigenetic X-chromosome state as a diagnostic tool for further refining the culture conditions of naive hPSCs.
Defining the in vivo X-chromosome states in mouse and human
In mammals, the presence of 2 X-chromosomes in females but one X- and one Y-chromosome in males creates a potential imbalance in gene dosage of X-linked and autosomal genes between males and females (3). X-chromosome inactivation (XCI) of one X in females is the canonical way, how mammals deal with this problem in somatic cells (4). It is mediated by the master regulator of XCI, the long non-coding RNA Xist (5-7), giving rise to one active and one inactive X-chromosome (XaXi). This differential X-chromosome state is associated with specific properties such as active chromatin configuration and X-linked gene expression from the Xa, and in contrast coating of the Xi by Xist RNA, which induces structural changes of the X-chromosome and leads to gene silencing and accumulation of epigenetic silencing marks such as DNA methylation and the histone modification H3K27me3 (8,9). Recent in vivo studies however (10,11) have shown differences in the mechanisms between mice and humans to achieve X-chromosome dosage compensation during preimplantation development (summarized in Figure 1). In mouse, first imprinted XCI is observed on the paternal X-chromosome, which gets progressively inactivated during preimplantation development. Then, X-chromosome reactivation (XCR) takes place in the epiblast of blastocysts at embryonic day (E)4.5; resulting in two active X-chromosomes (XaXa), while imprinted XCI is maintained in the extraembryonic primitive endoderm (PE) and trophectoderm (TE) lineages (5,12). XCR is followed by random XCI in the post-implantation epiblast, giving rise to one active and one inactive X-chromosome (XaXi) (4,13).
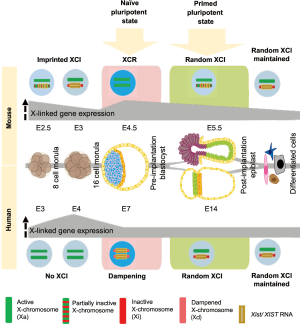
In humans however, recent in vivo studies of pre-implantation blastocysts showed drastic differences in X-chromosome state compared to mouse, based on RNA-FISH and allele-specific single-cell RNA-Seq analysis (2,10,11). Imprinted XCI does not occur during preimplantation development; instead, X-chromosome gene expression first increases during cleavage stages from E3 to E4 and then gradually decreases until E7 (blastocyst stage) in all three blastocyst cell lineages epiblast, PE and TE. Instead of imprinted XCI like in the mouse, where one X-chromosome is inactivated by Xist, human X-chromosome dosage compensation is achieved by lowering of gene expression from both X-chromosomes by half, defined as dampening of X-chromosome gene expression (10,11). The dampening mechanism is yet unclear, but also might involve XIST RNA, which is expressed from both X-chromosomes in this case. However, unlike during XCI, this does not result in accumulation of the H3K27me3 silencing mark. Therefore, we propose that in human females, X-chromosome activity in cells of the blastocyst could be represented as XdXIST+XdXIST+ (d stands for dampened) compared to XaXist−XaXist− (in epiblast) and XaXist−XiXist+ (in extraembryonic lineages) in mice. While mice undergo XCR in the epiblast in order to be able to switch from imprinted to random XCI, little is currently known about the transition from dampened X-dosage compensation to random XCI in the human post-implantation embryo and how this compares in terms of kinetics and mechanism to the mouse. Nevertheless, a partial shift from bi-allelic to mono-allelic X-linked gene expression in late human blastocysts has been observed, which might indicate the onset of that switch towards XCI (2).
The X-chromosome in mouse and human PSCs in vitro
The knowledge gained about the X-chromosome state from mouse and human embryos in vivo can serve as a blueprint, of what to expect from high quality pluripotent stem cells cultured in vitro. Epiblast cells from the inner cell mass (ICM) of blastocysts are the source of pluripotent embryonic stem cells (ESCs), which in mice reflect the naive or ground state of pluripotency thus called naive pluripotent cells (FBS/LIF or 2i/LIF culture conditions) (14,15). Post-implantation epiblast cells and their in vitro equivalent epiblast stem cells (EpiSCs) are in a so-called primed pluripotent state because they are primed for differentiation (FGF2/activin A culture conditions). In the mouse, differential X-chromosome states are indicators of these different cell potency states—XaXa for the naive pluripotent and XaXi for the primed pluripotent and differentiated cell states (Figure 1). However, in human the situation is a lot more complex since hESCs derived from pre-implantation blastocysts grown under conventional culture conditions similar to mouse EpiSCs (FGF2/activin A), do not display the X chromosome state of human pre-implantation epiblast cells in vivo (XdXd) and rather share molecular and functional properties with mouse post-implantation epiblast (14,15). For these so-called primed pluripotent hESCs, three distinct X-chromosome states have been described and categorized into classes (Table 1): class I as XaXIST−XaXIST− (same as mESCs), class II as XaXIST−XiXIST+ and class III as XaXIST−XeXIST− (Xe stands for an eroded Xi state, where XIST expression has been lost followed by a partial erosion of gene silencing accumulating over time) (16-19). As recapitulation of an in vivo-like naive pluripotent state is a major goal for human stem cell research and its potential clinical application, efforts have been made to convert primed hESCs to a naive-like state, both by transgene- and cytokine-mediated approaches using different culture conditions (see Table 2) (20-26). Based on the loss of XIST expression and the associated loss of H3K27me3 accumulation from the Xi, first studies suggested that naive hESCs reflect the mouse XaXa state thus being class I cells (20-24). However, as mentioned above, the majority of human blastocyst cells in vivo (~90% of total population) show two dampened X-chromosomes with bi-allelic XIST expression (XdXIST+XdXIST+), which we classify here as class IV cells (Table 1), while the remaining cells display various states from no to mono-allelic XIST expression (10,11). Therefore, if female human naive pluripotent stem cells were to reflect the in vivo situation regarding their X-chromosome status, they should be distinct from the class I mouse-like state (XaXIST−XaXIST−) but should rather be predominantly cells of class IV with bi-allelic XIST-RNA coating and a reduction of X-linked gene expression from both chromosomes by dampening (XdXIST+XdXIST+) (10,11). These fundamental differences regarding the X-chromosome state between mouse and human naive pluripotent cells in the blastocyst in vivo have redefined the gold standard, to which truly naive human ESCs in vitro should be compared. In that regard, two recent studies published in Cell Stem Cell provide for the first time a detailed characterization of the X-chromosome status of human naive ESCs (1,2).

Full table
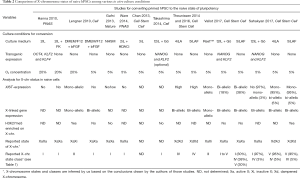
Full table
The X-chromosome in naive hPSCs—getting closer to in vivo
In the first study, Sahakyan et al. used 5iLAF (26) culture conditions to convert the primed female hESC line UCLA1 (class III, XaXIST−XeXIST−) into naive hESCs to examine, how the X-chromosome state changes during primed to naive conversion (Figure 2). By analyzing expression of X-linked genes and XIST passage by passage, they found that the conversion takes place through an intermediate state beginning around passage 4, where the cells would not show XIST expression and undergo XCR (similar to mESCs—class I XaXIST−XaXIST−). Similar to the situation in human pre-implantation blastocysts in vivo (27-29), autosomal and X-linked CpGs underwent progressive DNA-demethylation during the transition from the primed to the naive state. Interestingly, X-linked DNA-methylation was erased more rapidly than autosomal methylation marks, suggesting active DNA-demethylation during XCR being mechanistically distinct from the slower global demethylation. After passage 7 of culture in 5iLAF, XIST expression became upregulated, with cells expressing XIST mono-allelically from one X-chromosome being the dominant population (~95% of cells XaXIST−XaXIST+—in here we categorize them as class V cells) (Table 1). The prevalence of class V cells was also found by the second study on the subject by Vallot et al. (2) published in the same issue. Therefore, the most striking difference between human pre-implantation blastocysts and 5iLAF-cultured cells is the balance between class IV and class V cells: while the majority (~90%) of cells from blastocysts in vivo are of class IV (XdXIST+XdXIST+) with bi-allelic XIST expression, only 5% 5iLAF cultured hESCs reached the class IV “gold standard” as judged by RNA-FISH (28% when using more sensitive single-cell RNA-Seq). However, when hESCs were directly derived from human blastocysts under 5iLAF naive culture conditions, the proportion of class IV cells increased from 5% to 30%. XIST-upregulation in naive hESCs was accompanied by X-linked gene dosage compensation, when comparing X-linked gene expression levels between XIST-positive and XIST-negative naive hESCs. As X-linked genes were still bi-allelically expressed in XIST-positive cells, dosage compensation did not occur through X-inactivation but rather by dampening of expression, similar to the process observed in blastocysts in vivo (11). Interestingly, this bi-allelic X-dampening occurred even though XIST was predominantly mono-allelically expressed in naive hESCs. This raises the question, if and how XIST can play a direct role in the dampening process, as at least during X-inactivation, XIST acts solely in cis by coating and thereby silencing the X-chromosome from which it is expressed (8,9). Furthermore, despite reactivation of X-linked genes, naive UCLA1 cells cultured in 5iLAF displayed H3K27me3-enrichment on the XIST-expressing X-chromosome (1), differing from results of other studies (including Vallot et al.) of naive human cells in vitro and in vivo, where H3K27me3 was not enriched on the XIST-expressing X-chromosome(s) (2,10,20,23).
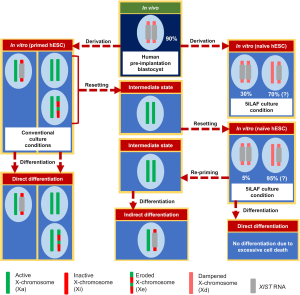
Taken together, these findings confirm that naive hESCs cultured in 5iLAF (and 4iLA) conditions recapitulate some of the characteristics attributed to human pre-implantation blastocyst cells, as they support the XIST-expressing XaXa state (mostly class V, XaXIST−XaXIST+). XCR has been also achieved under t2iL+Gö naive culture conditions (1,2,23,30), however XIST-expression was most consistently observed in 5iLAF (1,26,31). Additional improvements in culture conditions will still be necessary to fully recapitulate the in vivo X-chromosome state of the human blastocyst (class IV, XdXIST+XdXIST+) in hESC cultures. Furthermore, it will need to be clarified, if the dampening mechanism of X-linked gene expression is strictly XIST-dependent, since predominantly mono-allelic XIST expression can lead to bi-allelic dampening of X-linked gene expression in class V (XdXIST−XdXIST+) hESCs (1).
Epigenetic memory in naive hPSCs
Random XCI is a key characteristic of differentiated cells both in normal development and during in vitro culture. Thus, mESC differentiation in vitro recapitulates the changes X-chromosome state observed in vivo, resulting in a randomly selected active X and an Xist-expressing inactive X (XaXist−XiXist+). However, conventionally derived/primed hESCs after differentiation either don’t show XCI or maintain the XCI status of the original primed undifferentiated hESCs (16-18). In their study, Sahakyan et al. first tried to differentiate 5iLAF cultured naive UCLA1 cells directly but failed due to extensive cell death. Therefore, as an alternative strategy (Figure 2), they re-adapted naive cells before differentiation transiently to primed culture conditions (re-priming), resulting in two active X-chromosomes without XIST expression (XaXIST−XaXIST−, class I). Differentiation of naive cells via this intermediated stage showed XIST expression in about 80% of the cells with the majority of them undergoing XCI and XIST-mediated gene silencing, indicating that pre-priming is a critical step to faithfully achieve XCI when differentiating naive hESCs.
Interestingly, allele-specific expression analysis of primed, naive and re-differentiated hESC hiPSC lines displayed non-random XCI with the same X as in the original primed cells being inactivated upon differentiation (1). Thus, although XCR and transient XIST-downregulation occurred during establishment of naive hPSCs, the epigenetic memory of the previous X-inactivation state did not get erased, as it would normally occur in the germ cell lineage in vivo or in mouse pluripotent stem cells. This can be due to two reasons: Either the 5iLAF culture conditions are not yet optimized enough to capture the true epigenetic ground state in naive hESCs, which would include erasure of X-inactivation memory. Alternatively, the lack of X-inactivation memory erasure could reflect the absence of a need to erase imprinted XCI in the human blastocyst’s epiblast in contrast to the situation in mice since X-inactivation memory only needs to be erased in humans in the germ cell lineage in vivo. Additional studies on further optimizing naive hESC culture conditions could shed more light on this. Nevertheless, culture of epigenetically abnormal primed UCLA9 and UCLA4 hESCs of eroded Class III XCI-status in 5iLAF medium followed by differentiation via re-priming led to proper XCI, something which could not be achieved without the naive culture step (1,16-18). This shows that 5iLAF naive conditions are able to at least partially “cure” epigenetic abnormalities present in primed hESCs or hiPSCs. At least transient naive culture conditions could be therefore an important step when preparing high quality hPSCs for therapeutic applications.
XACT lncRNA—a marker for active and naive X-chromosome states
The study by Vallot et al. (2) focused predominantly on the role of the lncRNA XACT, which is linked to the active state of the human X-chromosome (32,33). In this paper, the authors show that XACT is co-expressed with similar kinetics to XIST from both X-chromosomes in human preimplantation embryos, suggesting initial transcriptional co-regulation of the two genes during early stages. After segregation of the three initial cell lineages epiblast, PE and TE, XIST and XACT became expressed more differentially, with biallelic co-expression in female epiblast and PE, however inconstant/decreasing XACT expression in TE, in which XIST remained bi-allelically expressed. XACT can form a RNA cloud similar to XIST and coats the single active X-chromosome in males and one or two X-chromosomes in female blastocysts. When co-expressed with XIST, the XACT RNA-domain is usually mutually exclusive with the XIST RNA-domain on the same X-chromosome with very little overlap. In female naive hESCs, XACT was consistently expressed from both X-chromosomes, mimicking the situation of pre-implantation epiblast in vivo. A previous study from the same group (32) suggested a potential role for XACT during erosion of X-chromosome silencing in primed hESCs, as XACT coating was an early mark of the eroding chromosome. Indeed, when XACT, which is a human-specific gene, was introduced as a transgene in mESCs on the mouse X-chromosome, Xist-upregulation after differentiation was biased towards the non-XACT-harbouring chromosome, suggesting that XACT might have an influence on X-inactivation choice in this transgenic context. It would be interesting to know, if XACT also plays a role in preventing of X-inactivation in hPSCs and early embryos, when it is co-expressed with XIST from the same X-chromosome and if this might contribute to X-chromosome dampening instead of XCI at these stages. Deletion studies of XACT on the human X-chromosome will be needed, to unequivocally demonstrate its role during human X-inactivation choice and/or in counteracting the silencing function of XIST in naive hESCs and early embryos.
Conclusions
The studies by Sahakyan et al. (summarized in Figure 2) and Vallot et al. (1,2) have significantly added to our understanding of the X-chromosome state in naive hESCs in vitro with respect to embryonic development in vivo. They have shown that 5iLAF culture conditions can convert primed to naive pluripotent hESCs with an X-chromosome state similar to pre-implantation blastocysts. The differences in dominant cell type between 5iLAF cultured naive cells (class V) and pre-implantation blastocysts (class IV) and the retention of epigenetic memory resulting in non-random XCI upon differentiation shows that further improvements of in vitro culture conditions will be necessary for better recapitulation of normal development. Furthermore, this work has now well proven that the X-chromosome state for naive hESCs is different from the one in mESCs; therefore, we believe the terms ‘naive’ and ‘primed’ as defined for mouse pluripotent stem cells cannot be used as a guideline to define hESCs anymore. Now, the naive pluripotent ground state reflecting the human pre-implantation blastocyst could be characterised as: presence of two active X-chromosomes expressing XIST with reduction of X-linked gene expression from both chromosomes by dampening (XdXIST+XdXIST+), absence of X-specific accumulation of the repressive histone modification mark H3K27me3, reduction in DNA-methylation and expression of the lncRNA XACT from one or both active X-chromosomes. Thus, the future in the field lies in further pursuit of in vitro culture conditions that mimic closely an X-chromosome state, which is found in the human pre-implantation blastocyst epiblast.
Acknowledgements
We would like to thank all members of the Payer lab for stimulating discussions.
Funding: SA Khan is funded by a Fellowship from the People Programme (Marie Curie Actions) of the European Union’s Seventh Framework Programme (FP7/2007-2013) under REA grant agreement n° 608959. We acknowledge support of the Spanish Ministry of Economy and Competitiveness, ‘Centro de Excelencia Severo Ochoa 2013-2017’, SEV-2012-0208 and for a Plan Estatal Grant to B Payer (BFU2014-55275-P). We also acknowledge the support of the CERCA Programme/Generalitat de Catalunya.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Sahakyan A, Kim R, Chronis C, et al. Human Naive Pluripotent Stem Cells Model X Chromosome Dampening and X Inactivation. Cell Stem Cell 2017;20:87-101. [Crossref] [PubMed]
- Vallot C, Patrat C, Collier AJ, et al. XACT Noncoding RNA Competes with XIST in the Control of X Chromosome Activity during Human Early Development. Cell Stem Cell 2017;20:102-11. [Crossref] [PubMed]
- Payer B, Lee JT. X chromosome dosage compensation: how mammals keep the balance. Annu Rev Genet 2008;42:733-72. [Crossref] [PubMed]
- Payer B. Developmental regulation of X-chromosome inactivation. Semin Cell Dev Biol 2016;56:88-99. [Crossref] [PubMed]
- Okamoto I, Otte AP, Allis CD, et al. Epigenetic dynamics of imprinted X inactivation during early mouse development. Science 2004;303:644-9. [Crossref] [PubMed]
- Namekawa SH, Payer B, Huynh KD, et al. Two-step imprinted X inactivation: repeat versus genic silencing in the mouse. Mol Cell Biol 2010;30:3187-205. [Crossref] [PubMed]
- Borensztein M, Syx L, Ancelin K, et al. Xist-dependent imprinted X inactivation and the early developmental consequences of its failure. Nat Struct Mol Biol 2017;24:226-33. [Crossref] [PubMed]
- Payer B. Epigenetic Regulation of X-Chromosome Inactivation. In: Naumova AK, Taketo T. editors. Epigenetics in Human Reproduction and Development. Singapore: World Scientific Publishing Co., 2017.
- da Rocha ST, Heard E. Novel players in X inactivation: insights into Xist-mediated gene silencing and chromosome conformation. Nat Struct Mol Biol 2017;24:197-204. [Crossref] [PubMed]
- Okamoto I, Patrat C, Thepot D, et al. Eutherian mammals use diverse strategies to initiate X-chromosome inactivation during development. Nature 2011;472:370-4. [Crossref] [PubMed]
- Petropoulos S, Edsgard D, Reinius B, et al. Single-Cell RNA-Seq Reveals Lineage and X Chromosome Dynamics in Human Preimplantation Embryos. Cell 2016;165:1012-26. [Crossref] [PubMed]
- Mak W, Nesterova TB, de Napoles M, et al. Reactivation of the paternal X chromosome in early mouse embryos. Science 2004;303:666-9. [Crossref] [PubMed]
- Lyon MF. Gene action in the X-chromosome of the mouse (Mus musculus L.). Nature 1961;190:372-3. [Crossref] [PubMed]
- Nichols J, Smith A. Naive and primed pluripotent states. Cell Stem Cell 2009;4:487-92. [Crossref] [PubMed]
- Weinberger L, Ayyash M, Novershtern N, et al. Dynamic stem cell states: naive to primed pluripotency in rodents and humans. Nat Rev Mol Cell Biol 2016;17:155-69. [Crossref] [PubMed]
- Mekhoubad S, Bock C, de Boer AS, et al. Erosion of dosage compensation impacts human iPSC disease modeling. Cell Stem Cell 2012;10:595-609. [Crossref] [PubMed]
- Patel S, Bonora G, Sahakyan A, et al. Human Embryonic Stem Cells Do Not Change Their X Inactivation Status during Differentiation. Cell Rep 2017;18:54-67. [Crossref] [PubMed]
- Silva SS, Rowntree RK, Mekhoubad S, et al. X-chromosome inactivation and epigenetic fluidity in human embryonic stem cells. Proc Natl Acad Sci U S A 2008;105:4820-5. [Crossref] [PubMed]
- Lessing D, Anguera MC, Lee JT. X chromosome inactivation and epigenetic responses to cellular reprogramming. Annu Rev Genomics Hum Genet 2013;14:85-110. [Crossref] [PubMed]
- Gafni O, Weinberger L, Mansour AA, et al. Derivation of novel human ground state naive pluripotent stem cells. Nature 2013;504:282-6. [Crossref] [PubMed]
- Hanna J, Cheng AW, Saha K, et al. Human embryonic stem cells with biological and epigenetic characteristics similar to those of mouse ESCs. Proc Natl Acad Sci U S A 2010;107:9222-7. [Crossref] [PubMed]
- Ware CB, Nelson AM, Mecham B, et al. Derivation of naive human embryonic stem cells. Proc Natl Acad Sci U S A 2014;111:4484-9. [Crossref] [PubMed]
- Takashima Y, Guo G, Loos R, et al. Resetting transcription factor control circuitry toward ground-state pluripotency in human. Cell 2014;158:1254-69. [Crossref] [PubMed]
- Lengner CJ, Gimelbrant AA, Erwin JA, et al. Derivation of pre-X inactivation human embryonic stem cells under physiological oxygen concentrations. Cell 2010;141:872-83. [Crossref] [PubMed]
- Chan YS, Goke J, Ng JH, et al. Induction of a human pluripotent state with distinct regulatory circuitry that resembles preimplantation epiblast. Cell Stem Cell 2013;13:663-75. [Crossref] [PubMed]
- Theunissen TW, Powell BE, Wang H, et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014;15:471-87. [Crossref] [PubMed]
- Pastor WA, Chen D, Liu W, et al. Naive Human Pluripotent Cells Feature a Methylation Landscape Devoid of Blastocyst or Germline Memory. Cell Stem Cell 2016;18:323-9. [Crossref] [PubMed]
- Okae H, Chiba H, Hiura H, et al. Genome-wide analysis of DNA methylation dynamics during early human development. PLoS Genet 2014;10:e1004868. [Crossref] [PubMed]
- Smith ZD, Chan MM, Humm KC, et al. DNA methylation dynamics of the human preimplantation embryo. Nature 2014;511:611-5. [Crossref] [PubMed]
- Collier AJ, Panula SP, Schell JP, et al. Comprehensive Cell Surface Protein Profiling Identifies Specific Markers of Human Naive and Primed Pluripotent States. Cell Stem Cell 2017;20:874-890.e7. [Crossref] [PubMed]
- Theunissen TW, Friedli M, He Y, et al. Molecular Criteria for Defining the Naive Human Pluripotent State. Cell Stem Cell 2016;19:502-15. [Crossref] [PubMed]
- Vallot C, Ouimette JF, Makhlouf M, et al. Erosion of X Chromosome Inactivation in Human Pluripotent Cells Initiates with XACT Coating and Depends on a Specific Heterochromatin Landscape. Cell Stem Cell 2015;16:533-46. [Crossref] [PubMed]
- Vallot C, Huret C, Lesecque Y, et al. XACT, a long noncoding transcript coating the active X chromosome in human pluripotent cells. Nat Genet 2013;45:239-41. [Crossref] [PubMed]
Cite this article as: Khan SA, Audergon PN, Payer B. X-chromosome activity in naive human pluripotent stem cells—are we there yet? Stem Cell Investig 2017;4:54.


