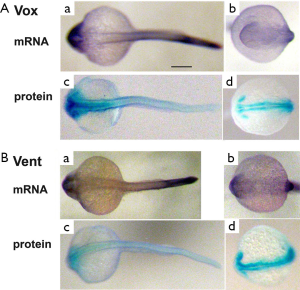
Expression of vox and vent mRNAs and encoded proteins in zebrafish embryos
Introduction
The vertebrate vent-family transcription factors play a crucial role in the establishment of the dorsoventral axis in embryos, by repressing organizer genes. Two zebrafish homeobox genes, vox/vega1 and vent/vega2, encode transcriptional repressors implicated in patterning of the dorsoventral axis (1-6). The vox also has an important function in regulation of ventral endoderm development and the separation of endoderm from mesoderm and ectoderm (7). Recently it was shown in medaka fish embryos (8), that vox was expressed in the developing neural tube similarly to central nervous system (CNS) markers. Thus the vent family genes, in addition to their fundamental role in body axis formation, may be involved in regionalization of vertebrate CNS.
A presence or absence of the specific mRNA in the cell is commonly considered as a presence or absence of the corresponding protein. However, there are lots of evidences that mRNA may be associated with inactive non-translated ribonucleoprotein complexes (9) and once synthesized protein may live long. Previously we have shown (10) that Xvent-2 mRNA in tails of Xenopus embryos was not active at the early tail bud stages. Although the Xvent-2 mRNA in Xenopus and vox mRNA in zebrafish embryos were detected in the tips of the tails, the Xvent-2 and vox proteins were revealed in other regions of the tails—along axial structures and around somites. In this study we for the first time compared the patterns of vent-family mRNAs and the encoded proteins in zebrafish embryos at early stages, before and after zygotic genes activation.
Methods
Embryo stages
Zebrafish embryos developmental stages were identified according to tables (11).
Isolation of RNA and RT-PCR analysis
Zebrafish embryos at stages 1–4 blastomeres were fixed with MEMFA solution (0.1 M MOPS pH 7.4; 2 mM EGTA; 2 mM MgSO4; formaldehyde 3.7%) for 6 hours at room temperature and 12 hours at 4 °C. Then they were transferred gradually into 70% ethanol and stored at −20 °C. RNA isolation was performed as in (12). The rehydrated embryos were washed sequentially with 2 M glycine containing 5 mM EDTA pH 8.0 and the buffer for proteinase K (10 mM Tris pH 8.0, 5 mM EDTA, 50 mM NaCl). To the last wash solution 1 mg of proteinase K and 30 mkg of glycogen were added. After incubation at 42 °C for 1 h, the sample was supplemented with 1 ml of the solution containing 40 mM Tris pH 7.5, 4% SDS, 0.4 M NaCl, and 10 mM EDTA and incubated for an additional 30 min at 42 °C and 30 min at 80 °C. Then the sample was transferred into a tube, and a standard phenol-chloroform deproteinization was carried on (13). After DNAse treatment the standard phenol–chloroform deproteinization was performed again and the isolated RNA was used in the reverse transcription reaction with the primer 5' TTCAGTAGTAATGATGTCTGGGCG 3'. The PCR was performed with this primer and the second primer 5' GAAATCATGGTGAAGAACTTTTCC 3'. The cDNA product was supposed to correspond to a full-length coding region of vox mRNA—736bp. The product of RT-PCR was analyzed by electrophoresis in 1.5% agarose gel.
Whole-mount in situ hybridization
Whole-mount in situ hybridization was carried out by a standard procedure (14). Plasmids pCS2+ carrying vent cDNA and pBS-SK carrying vox cDNA were used for the synthesis of digoxigenin-RNA by T3 RNA polymerase (Roche, Fermentas). The accession number for vox is AF255045 and for vent is AF255044. Plasmids were controlled by sequencing.
Microm HM 525 (Thermo Scientific) was employed for 18 µm cryosectioning.
Production of antibodies
Rabbit polyclonal antibodies against zebrafish vox and vent were produced by Almabion Company (Russia) using pvldvqepekktrphvpc and skfsvewlsqsfhdqekc peptides, respectively. Both antibodies were purified by affinity chromatography and conjugated with horseradish peroxidase. The same conjugate for rabbit polyclonal antibodies against human immunoglobulin (Imtek, Russia) was used in control experiments.
Western-blot analysis
The dechorionized embryos were homogenized in sample buffer and analyzed in Laemmli PAAG electrophoresis. Western-blot analysis was performed with single appropriate HRP-conjugated antibodies using SuperSignal Western Blot Enhancer Kit (Thermo Scientific, USA) according the manufacturer recommendations.
Whole-mount immunostaining
Embryos were primarily fixed in MEMFA solution, following procedures were described in (10).
Results and discussion
Expression of vox and vent mRNAs and the proteins before zygotic genes activation
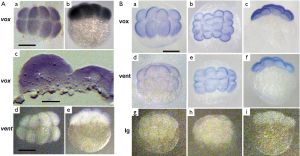
To know a spatio-temporal distribution of vox and vent mRNAs in zebrafish embryos, we performed the whole-mount in situ hybridization at different stages of embryonic development. We have shown, and somewhat reaffirmed the results of our previously published work (10), that the vox mRNA could be clearly revealed in embryos since the stage of 8–16 blastomeres [Figure 1A (a-c)]. Cryosectioning indicated that the cell nucleus were not stained and the vox mRNA staining was seen in cytoplasm, apparently associated with endoplasmic reticulum [Figure 1A (c)]. This result contradicts with the data, that the vox expression was activated immediately after midblastula transition (15). In contrast, the vent mRNA in our study was not revealed at early cleavage stages [Figure 1A (d, e)] like in (15). The whole-mount immunostaining revealed both vox and vent proteins at the stage of 8-16 blastomeres (Figure 1B).
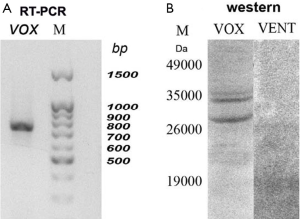
For validation, we had isolated the RNA from the earliest embryos (1–4 blastomeres) and performed the RT-PCR (Figure 2A). It is seen that the full-length coding vox mRNA was present in embryos. We supposed that by the TRIZOL method, as in (15) the RNA could not be extracted perfectly, but it was successfully extracted after proteinase treatment. Western blot analysis had also revealed vox and vent proteins at these stages (Figure 2B). It means that while the vox protein could be synthesized on the prestored transcripts or be prestored maternally, the vent protein was exclusively prestored maternally. The content of the vent protein in the embryos is very low, so the Western blot detected week bands of about 19 kDa. The vox showed quite good two bands. Such duplets were also described for some other proteins, for example (16), probably due to different extend of phosphorylation. Figure 2B also demonstrates an absence of cross-reaction with antibodies against vox and vent.
Expression of vox and vent mRNAs and the proteins after zygotic genes activation, at blastula, gastrula and segmentation periods
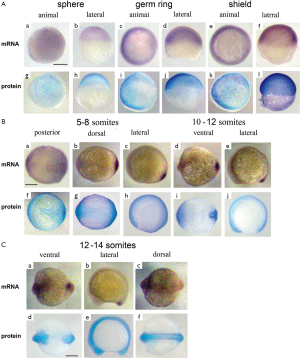
At the sphere stage the vox mRNA was expressed uniformly throughout the embryo except for a small region of clearing on the dorsal side [Figure 3A (a, b)]. The vox expression, by the shield stage, was heavy at the margin and cleared from a region on the dorsal side [Figure 3A (c-f)], corresponding to the organizer and neurectoderm. From the bud stage throughout somitogenesis, vox was expressed in the tailbud [Figure 3A (g, h); Figure 3B (a-f)], similar to other genes involved in the specification of ventral tissues (17,18). Up to the shield stage the vox protein staining progressively covered the embryos except for dorsal side [Figure 3A (i-p)], and resembled that of the mRNA. At the beginning of somitogenesis vox was clearly seen along the axis structures [Figure 3B (g-l) and Figure 3C (a-d)], while the mRNA at these stages was still concentrated in rostral and tail regions [Figure 3B (a-f)]. At the subsequent stages, the vox mRNA staining appeared along the dorsal side and the protein area covered that of mRNA and adjacent part of the yolk syncytial layer [Figure 3B (j-l)]. Earlier it was mentioned in literature that both vega 1 (vox) and vega 2 (vent) were also expressed within the yolk syncytial layer underlying the vega1 and vega2 expressing blastomeres (1,2).
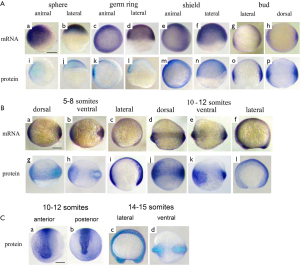
The vent mRNA was first expressed at the sphere stage [Figure 4A (a, b)], developing into a half-ring at the blastoderm margin, which became stronger by the shield stage. The vent expression at the shield stage [Figure 4A (c-f)] was confined to the margin, extending less far dorsally than the vox. Herewith the vent was mainly expressed posteriorly [Figure 4B (a-e); Figure 4C (a-c)] while the vox expression was extending from the tailbud to the rostral areas. The vent protein staining, since the sphere stage and up to the stage of bud [Figure 4A (g-l)], almost covered the mRNA staining area. From the bud stage throughout the somitogenesis, the vent mRNA staining was distinctly concentrated in the tail region while the protein staining was more extensive [Figure 4B (f-j); Figure 4C (d-f)].
Expression of vox and vent mRNAs and the proteins at the pharyngula period
The expression of vox mRNA and the protein at segmentation period was described in our previous work (19). At the pharyngula period stages the vox mRNA staining was concentrated mostly in rostral and distal tail areas, while the vox protein staining was revealed in the head region of the embryos and was absent from the tail tips (Figure 5A). The vent mRNA and the protein patterns slightly reminded those of the vox at these stages (Figure 5B). The vox and vent proteins were seen alongside the neural tube where neural crest cells were thought to be located, and also in presumptive eyes. Probably, the vox and vent regulate the activity of genes of the neural crest cells. This is consistent with our data on Xenopus embryos tails and zebrafish embryos at segmentation period (19).
Conclusions
Our results show that the spatial distributions of a protein and its mRNA may not always quite coincide during zebrafish embryogenesis. We observed such mismatches in embryos at the cleavage stage and in the pharyngula period. We propose that the cells, where a synthesis of a certain transcription factor had occurred, might migrate away from their native area.
Acknowledgements
We would like to thank Dr. D Onichtchouk for zebrafish embryos and plasmids kindly donated.
Funding: This study was supported by grant Russian Foundation for Basic Research No. 14-54 12008.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: All animal manipulations complied to European directive 2010/63, under the control of the veterinary services of Paris (authorization #75–419).
References
- Kawahara A, Wilm T, Solnica-Krezel L, et al. Antagonistic role of vega1 and bozozok/dharma homeobox genes in organizer formation. Proc Natl Acad Sci U S A 2000;97:12121-6. [Crossref] [PubMed]
- Kawahara A, Wilm T, Solnica-Krezel L, et al. Functional interaction of vega2 and goosecoid homeobox genes in zebrafish. Genesis 2000;28:58-67. [Crossref] [PubMed]
- Melby AE, Beach C, Mullins M, et al. Patterning the early zebrafish by the opposing actions of bozozok and vox/vent. Dev Biol 2000;224:275-85. [Crossref] [PubMed]
- Imai Y, Gates MA, Melby AE, et al. The homeobox genes vox and vent are redundant repressors of dorsal fates in zebrafish. Development 2001;128:2407-20. [PubMed]
- Ramel MC, Lekven AC. Repression of the vertebrate organizer by Wnt8 is mediated by Vent and Vox. Development 2004;131:3991-4000. [Crossref] [PubMed]
- Belting HG, Wendik B, Lunde K, et al. Pou5f1 contributes to dorsoventral patterning by positive regulation of vox and modulation of fgf8a expression. Dev Biol 2011;356:323-36. [Crossref] [PubMed]
- Zhao J, Lambert G, Meijer AH, et al. The transcription factor Vox represses endoderm development by interacting with Casanova and Pou2. Development 2013;140:1090-9. [Crossref] [PubMed]
- Fabian P, Pantzartzi CN, Kozmikova I, et al. vox homeobox gene: a novel regulator of midbrain-hindbrain boundary development in medaka fish? Dev Genes Evol 2016;226:99-107. [Crossref] [PubMed]
- Voronina AS, Pshennikova ES. mRNPs: From informosomes to stress granules. Mol Biol (Mosk) 2010;44:591-600. [Crossref] [PubMed]
- Voronina A, Pshennikova E. The Vox mRNA and protein expression in zebrafish Pou5f3 MZspg mutant embryos. Stem Cell Investig 2016;3:79. [Crossref] [PubMed]
- Kimmel CB, Ballard WW, Kimmel SR, et al. Stages of embryonic development of the zebrafish. Dev Dyn 1995;203:253-310. [Crossref] [PubMed]
- Voronina AS, Pshennikova ES. RNA isolation from the ribonucleoproteins fixed with formaldehyde. Prikl Biokhim Mikrobiol 2008;44:241-5. [PubMed]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Volume 2. Cold Spring Harbor Laboratory Press, 1989.
- Lehmann R, Tautz D. In situ hybridization to RNA. Methods Cell Biol 1994;44:575-98. [Crossref] [PubMed]
- Gilardelli CN, Pozzoli O, Sordino P, et al. Functional and hierarchical interactions among zebrafish vox/vent homeobox genes. Dev Dyn 2004;230:494-508. [Crossref] [PubMed]
- Lippok B, Song S, Driever W. Pou5f1 protein expression and posttranslational modification during early zebrafish development. Dev Dyn 2014;243:468-77. [Crossref] [PubMed]
- Connors SA, Trout J, Ekker M, et al. The role of tolloid/mini fin in dorsoventral pattern formation of the zebrafish embryo. Development 1999;126:3119-30. [PubMed]
- Joly JS, Joly C, Schulte-Merker S, et al. The ventral and posterior expression of the zebrafish homeobox gene eve1 is perturbed in dorsalized and mutant embryos. Development 1993;119:1261-75. [PubMed]
- Pshennikova ES, Voronina AS. The proteins of Vent-family and their mRNAs are located in different areas of the tails of Zebrafish and Xenopus embryos. Int J Biochem Cell Biol 2016;79:388-92. [Crossref] [PubMed]
Cite this article as: Pshennikova ES, Tereshina MB, Voronina AS. Expression of vox and vent mRNAs and encoded proteins in zebrafish embryos. Stem Cell Investig 2017;4:60.