
SETD7 at the heart of chromatin factor interplay
Cardiomyocyte (CM) differentiation is a complex, highly temporally and spatially controlled process that involves coordinated gene expression executed by transcription factors. These specific programs define the specification of mesodermal and cardiac lineages as well as the functioning of mature CMs. The stage specific transitions of transcriptional programs are also associated with epigenetic changes of DNA methylation, histone acetylation and methylation providing additional levels of control. Genome-wide epigenetic mark profiling studies unequivocally demonstrated the distinct patterns associated with each of the developmental changes (1).
The epigenetic enzyme regulation of the chromatin patterns in cardiac differentiation has been intensely investigated. New study in Cell Stem Cell highlights the role of lysine methyltransferase SETD7 in CM transcriptional programs (2). The authors’ observations on striking upregulation of SETD7 upon differentiation of human embryonic stem cell (hESC) to mesodermal progenitors (MES), CM progenitors (CP) and CMs has prompted in depth investigation of the function of this enzyme.
SETD7 is a protein lysine methyltransferase (PKMT) that monomethylates histone H3 lysine 4 (H3K4) as well as numerous other protein substrates. Wide variety of SETD7 functions has been described ranging from chromatin regulation and transcription, modulation of TP53, E2F1 driven apoptosis and proliferation to more specific signaling associated with methylation of YAP or ER transcription factors (3). Such wide array of functions and promiscuous substrate preference has both advanced and stymied our understanding of this PKMT.
The study by Lee and colleagues (2) takes a careful approach of characterizing SETD7 genomic localization in well-defined CM differentiation programs. Several SETD7 genome-wide binding patters were identified throughout ESC-MES-CP-CM differentiation that associate with functional lineage commitment transcriptional programs. Authors use knockouts and inducible knockdown of SETD7 at each of the differentiation stages to address the importance of SETD7 demonstrating the role in mesoderm and cardiac lineage commitment as well CM function. Elegant experiments identified the positive feedback between SETD7 and key regulator in the cardiac transcription factor network NKX2-5.
What enables SETD7 to drive the CM programs? To investigate the relationship of SETD7 and the epigenetic regulation, the authors investigated the association of SETD7 with activating/repressing marks as well as chromatin-associated co-factors and surprisingly found that although the co-occupancy exists for activator co-factors such as SWItch/Sucrose NonFermentable (SWI/SNF) family members and p300/CBP, SETD7 depletion or inhibition did not result in changes of total H3K4me1. Further analyses showed that SETD7 genomic binding sites are enriched for active chromatin marks such as H3K27ac, H3K4me3 and H3K36me3 (Figure 1).
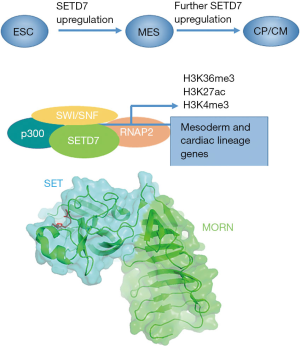
As the catalytic function of SETD7 did not seem to be necessary for cardiac program regulation, the possibility of scaffolding function was investigated performing the in vitro binding of H3K36 methylated and unmethylated peptides as SETD7 does not efficiently associate and methylate nucleosomes, possibly due to a relatively high surface electronegative charge (4). While these experiments pointed to possible association with histone tails, it is difficult to define the region of SETD7 that would be responsible for this function. Chromatin reader domains are usually distinctly separate and even though some PKMTs such as NSD enzymes both read and write chromatin marks, these functions are performed by separate domains (5). Thus, the question remains how SETD7 recognizes the chromatin and chromatin associated factors. SETD7 uniquely possesses N-terminal membrane occupation and recognition nexus (MORN) repeats that only are known for association with membrane phospholipids, however these repeats are essential for in vitro SETD7 activity (6) (Figure 1). Thus, more questions remain about this PKMT. It is also unclear how the essential function of SETD7 plays out in the case of the mouse knockout models where only mild phenotypes were noted (7). Several other PKMTs such as SMYD2 and SMYD1 have been linked to activating histone marks and CM biology thus it is possible that compensatory mechanisms exist. These wider questions await experimental answers.
Acknowledgments
Funding: The SGC is a registered charity (number 1097737) that receives funds from AbbVie, Bayer Pharma AG, Boehringer Ingelheim, Canada Foundation for Innovation, Eshelman Institute for Innovation, Genome Canada through Ontario Genomics Institute [OGI-055], Innovative Medicines Initiative (EU/EFPIA) [ULTRA-DD grant no. 115766], Janssen, Merck KGaA, Darmstadt, Germany, MSD, Novartis Pharma AG, Ontario Ministry of Research, Innovation and Science (MRIS), Pfizer, São Paulo Research Foundation-FAPESP, Takeda, and Wellcome [106169/ZZ14/Z].
Footnote
Conflicts of Interest: The author has no conflicts of interest to declare.
Ethical Statement: The author is accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Burridge PW, Sharma A, Wu JC. Genetic and Epigenetic Regulation of Human Cardiac Reprogramming and Differentiation in Regenerative Medicine. Annu Rev Genet 2015;49:461-84. [Crossref] [PubMed]
- Lee J, Shao NY, Paik DT, et al. SETD7 Drives Cardiac Lineage Commitment through Stage-Specific Transcriptional Activation. Cell Stem Cell 2018;22:428-44 e5.
- Batista IAA, Helguero LA. Biological processes and signal transduction pathways regulated by the protein methyltransferase SETD7 and their significance in cancer. Signal Transduct Target Ther 2018;3:19. [Crossref] [PubMed]
- Schapira M. Structural Chemistry of Human SET Domain Protein Methyltransferases. Curr Chem Genomics 2011;5:85-94. [Crossref] [PubMed]
- Arrowsmith CH, Bountra C, Fish PV, et al. Epigenetic protein families: a new frontier for drug discovery. Nat Rev Drug Discov 2012;11:384-400. [Crossref] [PubMed]
- Jacobs SA, Harp JM, Devarakonda S, et al. The active site of the SET domain is constructed on a knot. Nat Struct Biol 2002;9:833-8. [PubMed]
- Lehnertz B, Rogalski JC, Schulze FM, et al. p53-dependent transcription and tumor suppression are not affected in Set7/9-deficient mice. Mol Cell 2011;43:673-80. [Crossref] [PubMed]
Cite this article as: Barsyte-Lovejoy D. SETD7 at the heart of chromatin factor interplay. Stem Cell Investig 2019;6:20.


