
SETD7 in cardiomyocyte differentiation and cardiac function
Cellular differentiation is the process by which unspecialized cells mature into a variety of functional cell types to form the tissues of a multicellular organism. This specialization occurs during embryonic development when a single zygote gives rise to different cell types and in adulthood when reservoirs of stem cells differentiate to replace senescent or damaged cells. As cells differentiate, they undergo dramatic phenotypic changes that occur in parallel with temporal changes in gene expression as some genes become activated while others are silenced through epigenetic mechanisms. Embryonic stem (ES) cells have served as a powerful in vitro model system to decipher the function of epigenetic regulators in the process of differentiation as pluripotent stem cells, commit to a particular lineage, and then terminally differentiate into specialized cell types (1).
Epigenetic mechanisms that switch genes on and off during differentiation involve the activities of chromatin remodeling enzymes. These enzymes can be categorized as those, such as the switch/sucrose non-fermentable (SWI/SNF) type that utilize the energy from adenosine triphosphate (ATP) hydrolysis to disrupt chromatin structure or enzymes that add or remove post-translational modifications on histone proteins (2). SWI/SNF enzymes have been implicated in the differentiation of a broad array of cell types and function coordinately with enzymes that add or remove histone post-translational modifications in the regulation of gene expression (2). Histone post-translational modifications such as acetylation, phosphorylation, and methylation demarcate transcriptionally active or inactive chromatin. The enzymes that add these posttranslational modifications have traditionally been classified as “writers” while the enzymes that remove them as “erasers”. In addition, epigenetic “readers” play essential roles by interpreting chromatin modifications and recruiting effector proteins. Chromatin remodeling enzymes generally lack sequence specific binding ability and are recruited by interactions with transcription factors (TFs) and/or specific histone modifications (2). The dynamic and coordinated activities of each of these classes of chromatin remodeling enzymes result in a chromatin configuration that switches genes on or off during cellular differentiation. Interestingly, a recent study by Lee et al. in Cell Stem Cell suggests that the conventional role ascribed to the histone methyltransferase, SETD7, as an epigenetic “writer” is dispensable for cardiac differentiation, while a novel role as an epigenetic “reader” is essential (3).
SETD7 (also known as SET7/9, KIAA1717, KMT7) is a lysine methyltransferase (KMT) with a catalytic [Su(var)3-9, Enhancer of zeste and Trithorax] SET domain that can add methyl groups to histone H3K4 and linker histone H1 (4,5). The biological effects of its histone methylation activity have been enigmatic because SETD7 prefers free histones rather than histones assembled into nucleosomes (4). Furthermore, SETD7 methylates numerous non-histone proteins, including Suv39H1, p53, estrogen receptor alpha, DNMT1, E2F1, STAT3, RB, FXR, PARP1, YAP1, and PCAF and regulates transcription in a wide variety of biological processes (6). In contrast to these previous findings, Lee et al. found that SETD7 plays an essential role in cardiac differentiation that does not require its enzymatic activity. They carefully assessed the dynamic regulation of gene expression by SETD7 when human ES cells are differentiated into cardiomyocytes (CMs) and deciphered a novel role for SETD7 as an epigenetic reader of the H3K36me3 mark.
Lee et al. performed an in depth investigation into the role of SETD7 in the epigenetic regulation of cardiac differentiation. Utilizing a CM model of differentiation derived from human ES cells, they determined that SETD7 promotes stage-specific gene expression. When ESCs are induced to differentiate into CMs, they transition through a mesoderm (MES) cell stage (day 2), a cardiac progenitor (CP) cell stage (day 5) and then terminally differentiate into beating CMs (day 9). Cardiac specific genes began to be expressed on day 5 and reached maximum expression by day 9. SETD7 expression progressively increased throughout CM differentiation. Moreover, SETD7 expression also increased during murine CM differentiation and was highly expressed in adult human and baboon heart tissues. The observed induction of SETD7 expression prompted Lee et al. to investigate a potential role for SETD7 in CM differentiation and in maintenance of CM function.
Additional studies further supported a role for SETD7 in CM differentiation. Chromatin immunoprecipitation followed by high throughput sequencing (ChIP-seq) during CM differentiation demonstrated that SETD7 is dynamically associated with both promoters and gene bodies of actively transcribed genes that are required for cardiac development. Enrichment of SETD7 at these sites increased as the respective genes became activated and decreased as genes became silenced during the transition from ES to MES to CP to CM. Binding of SETD7 to pluripotency genes decreased during the differentiation process while binding to genes required for embryo patterning, somite and MES development as well as Notch and Wnt-signaling pathways transiently increased until the MES stage. After the MES stage, SETD7 was redistributed and detected primarily on genes required for cardiac development and heart morphogenesis. Thus, changes in SETD7 genome-wide binding were associated with gene expression changes that occur at multiple stages of CM differentiation.
Lee et al. then depleted SETD7 from ES cells using transcription activator-like effector nuclease (TALEN) and inducible shRNA approaches to determine the requirement for SETD7 at different stages of CM differentiation. Depletion of SETD7 at day 0 of differentiation suppressed MES lineage commitment and blocked cardiac lineage specification. There was a decrease in expression of key regulators required for MES commitment. These findings indicated that SETD7 is required early during CM differentiation for activation of MES-specific genes and failure to activate these genes prevents CM specification. Depletion of SETD7 just after MES commitment (day 3) and later in CPs (day 5) abrogated the expression of numerous cardiac specific TFs and cardiac structural genes. These findings are strong evidence that SETD7 is required for MES commitment, cardiac lineage specification, and terminal cardiac differentiation.
To elucidate the mechanisms by which SETD7 promotes commitment to the MES lineage, SETD7 interacting partners were identified at the MES stage. SETD7 was found to interact with p300 and the BAF60a and BRG1 components of the SWI/SNF chromatin remodeling complex. Importantly, SETD7 was required for recruitment of p300 and BRG1 to MES lineage promoters (Figure 1A). P300 is a histone acetyltransferase that deposits acetyl groups and interacts with the SWI/SNF complex to activate gene expression (7). BRG1, the catalytic subunit of the SWI/SNF complex, disrupts nucleosome structure and plays an important role in cardiac development, cardiac contractility, and cardiac hypertrophy (8-10). Because p300 and BRG1 increase chromatin accessibility, leading to induction of gene expression, these findings establish a mechanism by which SETD7 activates cardiac gene expression at the MES stage. However, the specific TFs which recruit SETD7 to MES lineage promoters were not identified in this study. Moreover, the interaction between BRG1 and SETD7 was not observed after the MES stage when BRG1 levels decrease, suggesting that SETD7 activates gene expression by different mechanisms in subsequent stages of CM differentiation.
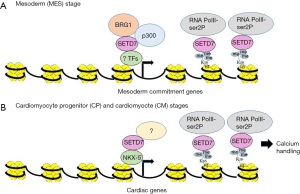
The requirement for SETD7 in later stages of CM differentiation relied heavily on its pivotal role in the cardiac specific transcriptional network. SETD7 was found to regulate genes encoding cardiac specific TFs, NKX2-5, TBX5, and GATA4 and to co-occupy NKX2-5, TBX5, and GATA4 consensus sites within cardiac specific target genes, indicating an extensive interplay between SETD7 and these cardiac specific TFs. Furthermore, SETD7 and NKX2-5 interacted in a positive feedback loop whereby NKX2-5 was bound to a consensus site on the SETD7 promoter and activated expression of SETD7 during CM differentiation and SETD7 occupied the NKX2-5 locus and was required for its expression. NKX2-5 physically interacted with SETD7 and was required to target SETD7 to a cardiac specific promoter. Thus, SETD7 is an essential component of the cardiac specific transcriptional network. However, the mechanisms by which SETD7 regulates transcription at the promoters of these genes were not determined. Does SETD7 activate transcription by recruiting a different cohort of co-factors which remodel chromatin and/or does it methylate histones or non-histone proteins at this stage of CM differentiation? Subsequent studies suggested that protein methylation was not a likely mechanism by which SETD7 activates transcription during CM differentiation.
As a KMT, SETD7 would be presumed to activate transcription of cardiac specific genes by methylating histone or non-histone proteins. Surprisingly, ES cells retained the ability to differentiate into CMs even when they were treated with PFI-2, a selective inhibitor of SETD7 enzymatic activity (6). Furthermore, PFI-2 treatment had no effect on MES or cardiac specific gene expression and did not affect global levels of H3K4me, suggesting that the methyltransferase activity of SETD7 is not required for expression of these genes nor for CM differentiation. These findings led the investigators to consider the possibility that SETD7 functions by a different epigenetic mechanism.
Cross-analysis of chip-seq data sets showed that genome-wide occupancy of SETD7 overlapped with H3K36me3 marks on gene bodies and coincided with highly expressed genes, suggesting a potential epigenetic cross-talk. Importantly, SETD7 co-occupied H3K36me3 enriched regions of cardiac specific promoters and gene bodies in differentiating CMs and in heart tissue. Further support for an association between SETD7 and H3K36me3 was seen by the high affinity of SETD7 for H3K36me3 on histone peptide arrays. SETD7 did not methylate histone peptides at H3K36 nor did depletion of SETD7affect H3K36me3 levels on its target genes, indicating that SETD7 does not deposit this epigenetic mark. However, H3K36me3 enrichment on cardiac specific genes was lost upon depletion of SETD2, a KMT that is known to catalyze H3K36 trimethylation (11). Loss of H3K36me3 by SETD2 depletion also disrupted SETD7 binding and compromised cardiac gene expression. These findings highlight a novel role for SETD7 as a reader of the H3K36me3 mark.
Epigenetic readers interpret the histone code by binding chromatin with specialized domains and then promoting transcription by various mechanisms. To elucidate the mechanisms by which SETD7 activates cardiac specific gene expression, a chip-seq experiment was conducted to investigate genome-wide changes in elongating RNA polymerase II occupancy as a function of SETD7 loss. Interestingly, association of RNA polymerase II, phosphorylated on serine 2, with the gene bodies of cardiac specific genes was disrupted in SETD7 knockout cells, primarily at H3K36me3 enriched regions. Since H3K36me3 has been associated with transcriptional activation and suppression of cryptic transcription sites, this finding indicated that SETD7 reads H3K36me3 marks to promote productive transcriptional elongation of cardiac specific genes (Figure 1B). Interestingly, although SETD7 does not possess a conserved Chromo, Tudor, or PWWP domain capable of reading the H3K36me3 mark, recombinant SETD7 protein was able to bind to H3K36me3 peptide arrays with high affinity, suggesting a direct interaction. Future studies that elucidate the regions of SETD7 which participate in this binding could uncover a novel motif or interacting partner that allows SETD7 to recognize the H3K36me3 mark in vivo.
SETD7 was also found to be required for proper CM function in terminally differentiated cells. High expression of SETD7 and co-occupancy with H3K36me3 was detected in both mature CMs and in adult heart tissue. Loss of SETD7 in terminally differentiated CMs caused arrhythmic beating and abnormal calcium handling. The expression of CASQ2, a major regulator of calcium storage and transport in CMs, was decreased as result of SETD7 depletion. SETD7 was found to bind to the CASQ2 gene body at H3K36me3 enriched regions, suggesting that SETD7 regulates CASQ2 expression by promoting transcriptional elongation. These molecular findings were consistent with an observed physiological effect of SETD7 depletion on CM contractility.
The finding that depletion of SETD7 disrupts normal CM beating suggests that SETD7 may be involved in cardiac arrhythmias. Interestingly, SETD7 regulates the expression of NKX2-5 and interacts with NKX2-5. Loss of NKX2-5 has previously been shown to result in cardiac arrhythmias through a mechanism that reactivates the NOTCH pathway in atrial CMs (12,13). Furthermore, polymorphisms near the NKX2-5 locus in humans have been correlated with heart rate abnormalities and predisposition to atrial fibrillation (14). It is intriguing that SETD7 was re-directed away from NOTCH loci as NKX2-5 expression increased after the MES stage. Additional studies could investigate how disruptions in the interactions between NKX2-5 and SETD7 impact upon SETD7 genome-wide binding and epigenetic regulation of gene expression in arrhythmias. These studies may illuminate new avenues to treat cardiac arrhythmias through epigenetic intervention.
Lee et al. also found that SETD7 is required for ES cell differentiation into neural, hepatocyte, and endothelial lineages. Together with previous studies that determined a requirement for SETD7 in pancreatic cell, smooth and skeletal muscle differentiation, Lee et al. demonstrated that SETD7 has an extensive role in cellular differentiation. However, it remains to be determined if SETD7 acts as a reader of H3K36me3 and whether SETD7 helps recruit components of the SWI/SNF complex to lineage specific promoters in these different lineages. Interestingly, like SETD7, BRG1 is also required for differentiation into a myriad of lineages (15). It is intriguing to speculate that SETD7 and BRG1 also interact to promote differentiation of other lineages at discrete stages of development.
Although Lee et al. provide strong data implicating SETD7 in cellular differentiation, previous studies reported that SETD7 knockout mice appear normal and do not have apparent developmental abnormalities (16). In contrast to the mouse studies, SETD7 disruption in zebrafish resulted in defective cardiac and skeletal muscle development and paralysis (17,18) and SETD7 disruption in Xenopus abrogated endoderm development to pancreas (19). Consistent with the Xenopus study, conditional disruption of SETD7 in the mouse pancreas interfered with beta cell function and resulted in glucose intolerance (20). Based on the extensive literature supporting a role for SETD7 in cellular differentiation in vitro and the findings by Lee et al., additional studies that more thoroughly investigate tissue function, particularly heart function in SETD7 knockout mice are warranted.
Emerging evidence suggests that SETD7 is involved in biological processes that impact upon numerous human diseases, including diabetes and other metabolic disorders, inflammation, fibrosis, and oncogenesis (21-23). Furthermore, studies in skeletal muscle suggest that SETD7 can be targeted for muscle regeneration (24) and for treating pulmonary fibrosis (25). Therefore, selective inhibition of SETD7 may have therapeutic potential. The study by Lee et al. highlights several novel interactions between SETD7, cardiac TFs, and BRG1 in cardiac development that can be further exploited therapeutically in cardiac diseases. Furthermore, the newly defined role of SETD7 as an epigenetic reader of H3K36me3 in heart now sets the stage for future studies of SETD7 in cardiac disease and the development of novel SETD7 small molecule inhibitors that target its reader function.
Acknowledgments
Funding: IL de la Serna was supported by funding from the National Institute of Arthritis and Musculoskeletal and Skin Diseases (ARO59379); the Melanoma Research Foundation; and a University of Toledo Innovation grant.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
References
- Keenen B, de la Serna IL. Chromatin remodeling in embryonic stem cells: regulating the balance between pluripotency and differentiation. J Cell Physiol 2009;219:1-7. [Crossref] [PubMed]
- Saladi SV, de la Serna IL. ATP dependent chromatin remodeling enzymes in embryonic stem cells. Stem Cell Rev 2010;6:62-73. [Crossref] [PubMed]
- Lee J, Shao NY, Paik DT, et al. SETD7 drives cardiac lineage commitment through stage-specific transcriptional activation. Cell Stem Cell 2018;22:428-44.e5. [Crossref] [PubMed]
- Wang H, Cao R, Xia L, et al. Purification and functional characterization of a histone H3-lysine 4-specific methyltransferase. Mol Cell 2001;8:1207-17. [Crossref] [PubMed]
- Castaño J, Morera C, Sesé B, et al. SETD7 regulates the differentiation of human embryonic stem cells. PLoS One 2016;11:e0149502. [Crossref] [PubMed]
- Barsyte-Lovejoy D, Li F, Oudhoff MJ, et al. (R)-PFI-2 is a potent and selective inhibitor of SETD7 methyltransferase activity in cells. Proc Natl Acad Sci U S A 2014;111:12853-8. [Crossref] [PubMed]
- Neish AS, Anderson SF, Schlegel BP, et al. Factors associated with the mammalian RNA polymerase II holoenzyme. Nucleic Acids Res 1998;26:847-53. [Crossref] [PubMed]
- Hang CT, Yang J, Han P, et al. Chromatin regulation by Brg1 underlies heart muscle development and disease. Nature 2010;466:62-7. [Crossref] [PubMed]
- Willis MS, Holley DW, Wang Z, et al. BRG1 and BRM function antagonistically with c-MYC in adult cardiomyocytes to regulate conduction and contractility. J Mol Cell Cardiol 2017;105:99-109. [Crossref] [PubMed]
- Mehta G, Kumarasamy S, Wu J, et al. MITF interacts with the SWI/SNF subunit, BRG1, to promote GATA4 expression in cardiac hypertrophy. J Mol Cell Cardiol 2015;88:101-10. [Crossref] [PubMed]
- Venkatesh S, Smolle M, Li H, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature 2012;489:452-5. [Crossref] [PubMed]
- Nakashima Y, Yanez DA, Touma M, et al. Nkx2-5 suppresses the proliferation of atrial myocytes and conduction system. Circ Res 2014;114:1103-13. [Crossref] [PubMed]
- Qiao Y, Lipovsky C, Hicks S, et al. Transient notch activation induces long-term gene expression changes leading to sick sinus syndrome in mice. Circ Res 2017;121:549-63. [Crossref] [PubMed]
- den Hoed M, Eijgelsheim M, Esko T, et al. Identification of heart rate-associated loci and their effects on cardiac conduction and rhythm disorders. Nat Genet 2013;45:621-31. [Crossref] [PubMed]
- de la Serna IL, Ohkawa Y, Imbalzano AN. Chromatin remodelling in mammalian differentiation: lessons from ATP-dependent remodellers. Nat Rev Genet 2006;7:461-73. [Crossref] [PubMed]
- Kurash JK, Lei H, Shen Q, et al. Methylation of p53 by Set7/9 mediates p53 acetylation and activity in vivo. Mol Cell 2008;29:392-400. [Crossref] [PubMed]
- Tao Y, Neppl RL, Huang ZP, et al. The histone methyltransferase Set7/9 promotes myoblast differentiation and myofibril assembly. J Cell Biol 2011;194:551-65. [Crossref] [PubMed]
- Kim JD, Kim E, Koun S, et al. Proper activity of histone H3 lysine 4 (H3K4) methyltransferase is required for morphogenesis during zebrafish cardiogenesis. Mol Cells 2015;38:580-6. [Crossref] [PubMed]
- Kofent J, Zhang J, Spagnoli FM. The histone methyltransferase Setd7 promotes pancreatic progenitor identity. Development 2016;143:3573-81. [Crossref] [PubMed]
- Maganti AV, Maier B, Tersey SA, et al. Transcriptional activity of the islet β cell factor Pdx1 is augmented by lysine methylation catalyzed by the methyltransferase Set7/9. J Biol Chem 2015;290:9812-22. [Crossref] [PubMed]
- Keating ST, El-Osta A. Transcriptional regulation by the Set7 lysine methyltransferase. Epigenetics 2013;8:361-72. [Crossref] [PubMed]
- Batista IAA, Helguero LA. Biological processes and signal transduction pathways regulated by the protein methyltransferase SETD7 and their significance in cancer. Signal Transduct Target Ther 2018;3:19. [Crossref] [PubMed]
- Elkouris M, Kontaki H, Stavropoulos A, et al. SET9-mediated regulation of TGF-β signaling links protein methylation to pulmonary fibrosis. Cell Rep 2016;15:2733-44. [Crossref] [PubMed]
- Judson RN, Quarta M, Oudhoff MJ, et al. Inhibition of methyltransferase Setd7 allows the in vitro expansion of myogenic stem cells with improved therapeutic potential. Cell Stem Cell 2018;22:177-90.e7. [Crossref] [PubMed]
- Tamura R, Doi S, Nakashima A, et al. Inhibition of the H3K4 methyltransferase SET7/9 ameliorates peritoneal fibrosis. PLoS One 2018;13:e0196844. [Crossref] [PubMed]
Cite this article as: Basuroy T, de la Serna IL. SETD7 in cardiomyocyte differentiation and cardiac function. Stem Cell Investig 2019;6:29.


