
Clocking the circadian genes in human embryonic stem cells
Introduction
All living organisms respond to changing environmental conditions, as well as to predictable cyclic conditions such as light-dark cycles, high-low tides and summer-winters. To cope with cyclical changing conditions, all organism have evolved ways to deal with the changing conditions, and this requires precise control over gene expression. The circadian cycle is a mechanism that enables animals to perform important functions such as sleep, metabolism, immune function, tissue regeneration, memory formation, differentiation, hormone secretion, blood pressure control, body temperature among others (1-6). It has been shown that light is an important inducer of circadian cycle in animals as well as plants. At the molecular level, the circadian system works due to cyclical expression of transcription factors such as BMAL-1, CLOCK, PERIOD 1–3 (PER1–3), CRPTOCHROME 1–2 (CRY1–2), REV-ERα and REV-ERβ. BMAL-1 and CLOCK form a heterodimer, that activates the transcription of PERIOD, CRYPTOCHROME, REV-ERα and REV-ERβ genes, these in turn lead to repression of BMAL1 and CLOCK genes (3,7). However, PER, CRY, REV-ERα and REV-ERβ proteins have shorter half-life and are destroyed, which relieves the repression of BMAL-1 and CLOCK genes, again the cycle restarts from BMAL1: CLOCK expression, this happens in a cyclical manner (3).
Knockout studies of various circadian genes in mice have helped understand the role of the circadian cycle in normal development. Bmal1 knockout mice are infertile, have impaired glucose regulation, show accelerated ageing, reduced bone and muscle mass (8). Mice with Clock and Period gene knockouts show hyperphagia and diet induced obesity and they also developed various lipid disorders under different dietary conditions (9). Mice with knockout of Clock gene are normal at birth, but they have reduced life span, cataracts and persistent skin inflammation (10). The knockout studies of Cry gene have shown that it plays essential role in maintaining energy homeostasis. Cryptochrome knockout mice studies showed that, these mice have a normal circadian cycle when exposed to a 12-hour light/dark cycle, but the double mutant mice showed increased insulin secretion that leads to excessive adipose tissue deposition (11). Data on mutations in core circadian genes in human diseases is restricted mostly to neuropsychiatric disorders.
Most cells in our body follow a circadian rhythm, whereas in case of transplanted if they do not sync with the host’s circadian rhythm, the graft may not function optimally. Molecular analysis of various circadian genes in different mouse organs such as liver, adrenal gland, brainstem, heart, hypothalamus, showed that circadian gene expression varied widely among the different organs with the highest in mouse liver cells (12) There are several clinical trials involving use of human pluripotent stem cell derived functional cells (13), and it would be important to find out if they can sync their gene expression post transplantation. We studied the expression of circadian genes such as CLOCK, BMAL-1, CRYPTOCHROME and PERIOD in human embryonic stem cells in undifferentiated state and spontaneously differentiated cells; and found that human pluripotent stem cells show cyclical expression of circadian genes.
Methodology
Cell culture
Human embryonic stem cell line KIND1, was procured from National Institute for Research in Reproductive Health (NIRRH). For culturing KIND 1 cells, culture dishes were coated with 1X Vitronectin (Thermo Scientific, CA, USA) for 1 hour at 37 °C in DPBS and then KIND1 cells were grown in Essential 8 medium (Thermo Scientific). KIND1 cells showing >80% confluency were passaged using 10 mM EDTA (Sigma Aldrich, MO, USA). The undifferentiated cells were harvested at day 4 (at this stage the cells show peak confluency), and subsequently at 6, 18 and 22 hours, with media changes performed daily. To induce differentiation, the undifferentiated cells on day 4 were first given a wash with DPBS and then DMEM containing 10% fetal bovine serum (FBS, Thermo Scientific) was added to the cells. The cells were allowed to differentiate at 37 °C and 5% CO2 humidified atmosphere and harvested at 6, 18 and 22 hours. KIND1 cells were imaged at 10× magnification using brightfield microscope (AxioCam ERc 5s, Carl Zeiss, Germany). The cells were harvested for RNA extraction at 6, 18 and 22 hours, post seeding of undifferentiated cells or post induction of differentiation.
Primer design
Primers were designed using Primer Blast https://www.ncbi.nlm.nih.gov/tools/primer-blast/. The annealing temperature for primers were standardized using mixture of differentiated and undifferentiated human pluripotent stem cell cDNA. Sequences for OCT4, NANOG, SOX2, CLOCK, BMAL-1, PERIOD, CRYPTOCHROME and β2-MICROGLOBULIN are given in Table 1.
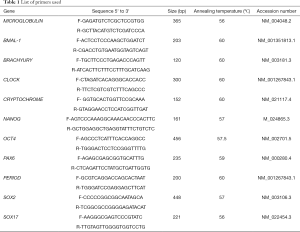
Full table
Reverse transcription polymerase chain reaction
Total RNA in differentiated and undifferentiated KIND1 cells at various time points was extracted using TRI reagent (Sigma Aldrich) as per manufacturer’s instructions. The RNA was quantitated using microplate reader (Gen5 BioTek Instruments Inc., VT, USA) and the total RNA concentration was estimated from its absorbance at 260 nm. Primescript 1st strand cDNA synthesis kit (TakaraBio, Kusatsu, Japan) was used to convert 1 µg of total RNA into cDNA, and this conversion was done as per manufacturer’s instructions using ABI Thermal Cycle (Applied Biosystems, CA, USA) in a 20 µL reaction volume. Emerald Green 2X PCR mix (TakaraBio) was used for performing the PCR and done as per manufacturer’s instructions using ABI Thermal Cycle (Applied Biosystems, CA, USA) in a 25 µL reaction volume. The cycling parameters for PCR reaction were as follows: initial denaturation at 94 °C for 3 min, denaturation at 94 °C for 30 s, annealing (as given in Table 1) for 30 s, elongation at 72 °C for 30 s these three cycles were repeated 34 times, with 7 min final extension at 72 °C. The PCR products were separated by performing electrophoresis using 2% agarose with 0.5 µg/mL ethidium bromide (Bangalore Genei, Bangalore, India) was used for detecting PCR products. The 100 bp DNA ladder (Thermo Scientific) was used for determine the size of the PCR products on agarose gels. The ethidium bromide stained gels were imaged in the ChemiDoc gel documentation system (Bio-Rad, IL, USA) and the images obtained were quantitated using Image J software (http://rsbweb.nih.gov/ij) and using β2-MICROGLOBULIN for normalizing.
Results
Differentiation of human pluripotent stem cells
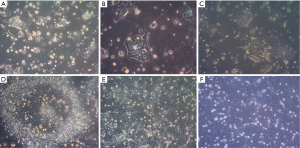
Human embryonic stem cell line KIND1, at 70% confluency was seeded onto vitronectin coated 35-mm dish. At 6 hours the cells attach to the dish as small cell clusters of 3 to 4 cells (Figure 1A), and later the cells possibly undergo one cell cycle (Figure 1B,C). Simultaneously, in another set of 35-mm dishes, KIND1 cells were allowed to reach 70% confluency and at day 4, they were differentiated using DMEM F12 supplemented with 10% FBS. Once the cells are induced to differentiate, they appear flattened and larger at 6, 18 and 22 hours (Figure 1D,E,F). The expression of pluripotency controlling genes OCT4, NANOG and SOX2 was seen using RT-PCR and Figure 2A shows the cells expressed OCT4, NANOG and SOX2, however no expression of lineage specific genes was seen (data not shown) in the cells grown with Essential 8 medium. We chose three representative genes—BRACHYURY, PAX6 and SOX17 to access spontaneous differentiation into mesoderm, ectoderm and endoderm lineages respectively. When KIND1 cells were induced to differentiate using media containing 10% FBS (final concentration), expression of BRACHYURY (mesoderm specific) and PAX6 (ectoderm specific) genes was seen from 6 hours onwards (Figure 2B). However, no expression of early endoderm specific transcription factor SOX17 was seen. Thus, upon differentiation cells primarily of mesoderm and ectoderm would have formed by the end of 22 hours, but the bands for BRACHURY and PAX6 appear faint, this could be due to the short time frame we used, if we had extended the time to 48 hours probably robust expression of lineage specific markers would have been seen.
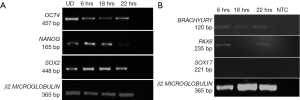
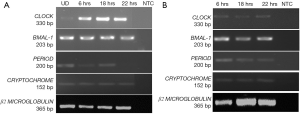
Expression of circadian gene in human pluripotent stem cells
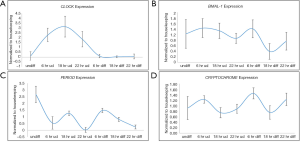
Once, we established that we had both undifferentiated and differentiated human embryonic stem cells, we moved ahead to determine expression of the core circadian genes. In order to understand the circadian cycle gene expression in undifferentiated human pluripotent stem cells, we harvested RNA at 6, 18 and 22 hours after passaging, as well as cells on the eve of passaging (days 4–5) to serve as control. For the initial 24 hours after seeding the human embryonic stem cells appear slightly mesenchymal like in morphology (Figure 2A), but as the cells proliferate, they attain epithelial morphology. We performed RT-PCR for genes associated with pluripotency—OCT4, NANOG and SOX2, and observed that all three genes were expressed at all the time points (Figure 2A). Once we established that KIND1 cells were pluripotent, we then carried out RT-PCR to investigate expression of circadian genes namely CLOCK, BMAL-1, PERIOD and CRYPTOCHROME. We found that PERIOD, BMAL-1 and CRYPTOCHROME genes were expressed at 6, 18 and 22 hours post seeding, however CLOCK gene expression was lowest in day 4 undifferentiated cells but was highly expressed post seeding (Figure 3A). We then looked for expression of CLOCK, BMAL-1, PERIOD and CRYPTOCHROME at 6, 18 and 22 hours post addition of differentiation media (Figure 3A,B). Interestingly expression of CLOCK was much lower in differentiated human embryonic stem cells, and PERIOD which was absent at 22 hours in undifferentiated cells was expressed at all time points in differentiated cells (Figure 3A,B). Using several agarose gels images (n=4) for every time point, we used ImageJ to perform semi-quantitative analysis. We observed that CLOCK expression seems to be downregulated in differentiated cells (Figure 4A), while BMAL-1, PERIOD and CRYTOCHOME show cyclicity (Figure 4B,C,D).
Discussion
Our work demonstrates that core circadian cycle genes CLOCK, BMAL-1, PERIOD and CYRPTOCHROME shown cyclicity in human pluripotent stem cells in undifferentiated state as well as upon spontaneous differentiation into mesoderm and ectoderm, even though there are no definite light/dark cycles for the in vitro cultured cells. After decades of painstaking work, the set of neurons in the brain called the superchiasmatic nuclei were found to control the circadian rhythm (14). The circadian proteins along with chromatin modifiers, RNA binding proteins, phosphatases and kinases control expression of genes in a rhythmic manner (15-17). Human pluripotent stem cells have the ability to differentiate into any cell lineage and they can be used in cell replacement therapies, but if circadian genes play pivotal role in the differentiation process, then it is imperative to understand their dynamics in human pluripotent stem cells. Circadian genes are crucial for proliferation and functioning of intestinal cells, liver cells, hormone producing cells, heart cells and immune cells (2,12,18,19) however their expression pattern in human pluripotent stem cells has not been completely elucidated.
Bmal-1 has been shown to control expression of inflammatory cytokines that are required for proliferation of intestinal cells, and mice with mutated Bmal-1 show impaired intestinal regeneration (18). Rogers et al. 2017 compared expression of Period, Bmal-1 and Rev ErbAα in human bone marrow derived stem cells, adipose-derived stem cells and dental-derived stem cells (20). They showed differential expression of Period, Bmal-1 and Rev ErbAα in different human adult stem cells, however authors appeared to have shown normalized expression but qRT-PCR inverse normalized expression should be given. Recently, it was shown that by simply adjusting cells to regular feeding and fasting cycles, the in vitro generated islet cells get entrained and these cells become functionally mature islets cells in vitro (21).
Dierickx et al. 2017, using human embryonic stem cells showed that functional circadian genes are expressed but no rhythmicity was seen when they were in undifferentiated state, however once they were differentiated into cardiomyocytes the circadian genes showed rhythmicity (19). We also, show that undifferentiated human embryonic stem cells show expression of core circadian genes, but the difference in cyclicity could be due to shorter time point we used. In mouse embryonic stem cells, the circadian genes do not oscillate, however upon differentiation oscillation in circadian genes was seen and interestingly when the differentiated cells were reprogrammed using OCT4, SOX2, KLF4 and c-MYC (OSKM), the circadian gene oscillation ceased (22). Our results show that human embryonic stem cells and its differentiated progeny both show circadian gene oscillations. However, it would be crucial to determine what are the molecular regulators of CLOCK, BMAL1, PERIOD and CRYPTOCHOME. Most studies have shown that the circadian genes are critical in differentiation or for functionality of the differentiated cells, however the function of circadian gene in undifferentiated human embryonic stem cells needs to be investigated. It has been established that light exposure keeps the cyclical expression of the circadian genes, however in vitro differentiated cells are not exposed to light rhythmically, yet we find expression of the circadian genes, which opens up possibilities for chemical inducers of circadian genes and it merits further investigations.
It would be indeed interesting to determine how cyclical oscillations of circadian genes in human pluripotent stem cells in vitro are regulated, and results from Alvarez-Dominguez et al. 2020, have shown that if we can control the circadian genes it can greatly affect the functionality of cells generated from human pluripotent stem cells (21).
Conclusions
Human embryonic stem cells exhibit circadian oscillations in vitro both in undifferentiated and spontaneously differentiated cells. Human embryonic stem cells show expression of core circadian gene such as CLOCK, BMAL1, PERIOD and CYRPTOCHROME.
Acknowledgments
The authors thank NMIMS Sunandan Divatia School of Science for providing the necessary facilities, reagents and encouragement to undergraduate students to carry out the research work.
Funding: None.
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/sci-2020-014). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Cedernaes J, Osler ME, Voisin S, et al. Acute Sleep Loss Induces Tissue-Specific Epigenetic and Transcriptional Alterations to Circadian Clock Genes in Men. J Clin Endocrinol Metab 2015;100:E1255-61. [Crossref] [PubMed]
- Cao Q, Zhao X, Bai J, et al. Circadian clock cryptochrome proteins regulate autoimmunity. Proc Natl Acad Sci U S A 2017;114:12548-53. [Crossref] [PubMed]
- Dierickx P, Van Laake LW, Geijsen N. Circadian clocks: from stem cells to tissue homeostasis and regeneration. EMBO Rep 2018;19:18-28. [Crossref] [PubMed]
- Tsang AH, Astiz M, Friedrichs M, et al. Endocrine regulation of circadian physiology. J Endocrinol 2016;230:R1-11. [Crossref] [PubMed]
- Douma LG, Gumz ML. Circadian Clock-Mediated Regulation of Blood Pressure. Free Radic Biol Med 2018;119:108-14. [Crossref] [PubMed]
- Gamble KL, Berry R, Frank SJ, et al. Circadian Clock Control of Endocrine Factors. Nat Rev Endocrinol 2014;10:466-75. [Crossref] [PubMed]
- Lande-Diner L, Boyault C, Kim JY, et al. A positive feedback loop links circadian clock factor CLOCK-BMAL1 to the basic transcriptional machinery. Proc Natl Acad Sci U S A 2013;110:16021-6. [Crossref] [PubMed]
- Yang G, Chen L, Grant GR, et al. Timing of expression of the core clock gene Bmal1 influences its effects on aging and survival. Sci Transl Med 2016;8:324ra16. [Crossref] [PubMed]
- Tsang AH, Astiz M, Leinweber B, et al. Rodent Models for the Analysis of Tissue Clock Function in Metabolic Rhythms Research. Front Endocrinol (Lausanne) 2017;8:27. [Crossref] [PubMed]
- Dubrovsky YV, Samsa WE, Kondratov RV. Deficiency of circadian protein CLOCK reduces lifespan and increases age-related cataract development in mice. Aging (Albany NY) 2010;2:936-44. [Crossref] [PubMed]
- Barclay JL, Shostak A, Leliavski A, et al. High-fat diet-induced hyperinsulinemia and tissue-specific insulin resistance in Cry-deficient mice. Am J Physiol Endocrinol Metab 2013;304:E1053-63. [Crossref] [PubMed]
- Zhang R, Lahens NF, Balance HI, et al. A circadian gene expression atlas in mammals: Implications for biology and medicine. Proc Natl Acad Sci U S A 2014;111:16219-24. [Crossref] [PubMed]
- Eguizabal C, Aran B, Chuva de Sousa Lopes SM, et al. Two decades of embryonic stem cells: a historical overview. Hum Reprod Open 2019;2019:hoy024.
- Stephan FK, Zucker I. Circadian rhythms in drinking behavior and locomotor activity of rats are eliminated by hypothalamic lesions. Proc Natl Acad Sci U S A 1972;69:1583-6. [Crossref] [PubMed]
- Brown SA. Circadian clock-mediated control of stem cell division and differentiation: beyond night and day. Development 2014;141:3105-11. [Crossref] [PubMed]
- Jagannath A, Taylor L, Wakaf Z, et al. The genetics of circadian rhythms, sleep and health. Hum Mol Genet 2017;26:R128-38. [Crossref] [PubMed]
- Weger M, Diotel N, Dorsemans AC, et al. Stem cells and the circadian clock. Dev Biol 2017;431:111-23. [Crossref] [PubMed]
- Stokes K, Cooke A, Chang H, et al. The Circadian Clock Gene BMAL1 Coordinates Intestinal Regeneration. Cell Mol Gastroenterol Hepatol 2017;4:95-114. [Crossref] [PubMed]
- Dierickx P, Vermunt MW, Muraro MJ, et al. Circadian networks in human embryonic stem cell-derived cardiomyocytes. EMBO Rep 2017;18:1199-212. [Crossref] [PubMed]
- Rogers EH, Fawcett SA, Pekovic-Vaughan V, et al. Comparing Circadian Dynamics in Primary Derived Stem Cells from Different Sources of Human Adult Tissue. Stem Cells Int 2017;2017:2057168.
- Alvarez-Dominguez JR, Donaghey J, Rasouli N, et al. Circadian Entrainment Triggers Maturation of Human In Vitro Islets. Cell Stem Cell 2020;26:108-122.e10. [Crossref] [PubMed]
- Yagita K, Horie K, Koinuma S, et al. Development of the circadian oscillator during differentiation of mouse embryonic stem cells in vitro. Proc Natl Acad Sci U S A 2010;107:3846-51. [Crossref] [PubMed]
Cite this article as: Thakur S, Storewala P, Basak U, Jalan N, Pethe P. Clocking the circadian genes in human embryonic stem cells. Stem Cell Investig 2020;7:9.


