
Tamoxifen induces stem-like phenotypes and multidrug resistance by altering epigenetic regulators in ERα+ breast cancer cells
Introduction
Breast cancer is the most common cancer in women and the leading cause of cancer-related deaths worldwide (1). In the past decade, the incidence of breast cancer was increasing significantly among younger women population. About 70% of the breast carcinomas were estrogen receptor alpha positive [ERα+] (2,3). Selective estrogen receptor modulators (SERMs), such as 4-hydroxy-tamoxifen (TAM) and raloxifene, play an important role in the treatment of ERα+ breast tumors. In addition, the SERMs improve the treatment potentially in the early stages of breast cancer. Eventually, the treatment with TAM resulte in an increase in the development of endocrine-resistant breast tumor phenotypes that lead to the formation of secondary tumors and multidrug resistance (4). Approximately 50% of ERα+ breast tumors are metastatic and do not respond to endocrine therapies. Around 40% of early stage breast tumors that are treated with endocrine therapies develop endocrine resistance. Previous studies (5,6) have shown that the treatment with TAM induces stem-like phenotypes and develop multidrug resistance, which are associated with the enhanced expression of growth factor receptors, such as EGFR and HER2. Therefore, it is important to study the endocrine resistance by using ERα+ breast cancer cell models.
MCF-7 cell line expresses ERα+/HER2− (7). The proliferation and survival of MCF-7 cells are dependent on estrogens that activate estrogen responsive genes and this cell line has been used widely to demonstrate endocrine resistance (8,9). Tamoxifen resistance (TR) associated with the development of multidrug resistance (MDR) has been considered as an important phenomenon in the breast cancer cells (10,11). TR-associated development of MDR has been a major obstacle in the treatment of breast cancer and is associated with the overexpression of ATP-binding cassette-binding proteins, such as P-glycoprotein (P-gp/ABCB1), Breast Cancer Resistant Protein (BCRP/ABCG2) and Multidrug Resistant Protein-1 (MRP-1/ABCC1) (4,12). Previous studies (13,14) have shown that TAM treatment is associated with the development of MDR in breast cancer cells.
The treatment using TAM resulted in the chronic epigenetic changes, such as DNA methylation, histone methylation, demethylation, acetylation, phosphorylation and sumoylation associated with TR and MDR (15-17). Previous studies (18-20) have reported that histone H3 lysine 4-specific methyltransferase (SETD1A) and H3 histone lysine demethylase 3A (KDM3A) are overexpressed in TR breast cancer cells. In a recent study (21), it was reported that an elevated expression of KDM3A leads to an increase in the development of pancreatic cancers. However, the mechanisms through which TAM induces MDR and stem-like phenotypes in breast cancer cells are unclear. Moreover, epigenetic alteration is an important phenomenon to study the development of stem-like phenotypes and MDR. Unfortunately, only a few studies are available with regard to the development of MDR and stem-like phenotypes in breast cancer cells.
Therefore, we determined the TAM-induced epigenetic changes associated with stem-like changes and the development of MDR in breast cancer cells. The treatment with TAM enhanced Notch-1 gene expression, overexpression of MDR genes and stem cell marker genes in the breast cancer cells.
Methods
Fetal bovine serum (FBS) and phenol red free DMEM were obtained from HiMedia (India). HEPES, trypan blue, sodium pyruvate, and sodium bicarbonate were purchased from Sisco Research Laboratories (Mumbai, India). Crystal violet, bovine serum albumin, and water-soluble 17β-estradiol, dextran-sulfate T70, 4-hydroxy-tamoxifen, 3[4,5-Dimethylthiazol-2-yl]-2,5-diphenylterazolium bromide (MTT) and doxorubicin were obtained from Sigma-Aldrich (USA). The anti-human Notch-1 primary antibody (Santa Cruz Biotechnology, USA), anti-human CD44 (Thermo Scientific, USA), β-actin (Sigma-Aldrich), anti-mouse HRP and anti-rabbit HRP (Santa Cruz Biotechnology, USA) were obtained. Dynabead magnetic immunoprecipitation kit derived from Thermofisher (USA). QRT-PCR array and master mix were obtained from Qiagen (Germany). ALDEFLOUR assay kit, MammoCultTM media and ultralow attachment plates were obtained from Stemcell Technologies (Canada).
Cell culture
MCF-7 cell line was procured from NCCS (Pune, India). The cells were cultured with and without phenol red media. The cells were supplemented with 10% fetal bovine serum (FBS), 1% penicillin/streptomycin and cultured in 5% CO2 at 37 °C in a CO2 incubator. Once the cells attained confluence, the cells were treated with trypsin-EDTA, subcultured and plated at a density of 1×104 cells in 96-well plates to perform cell viability assay. Cells were cultured in 6-well plates for mammosphere formation assay and cultured in 10 cm2 plates for western blot analysis.
Charcoal-stripped serum (CSS) preparation
The 1 nmol/L HEPES buffer (pH 7.4) was prepared and 5% activated charcoal (w/v) and Dextran T70 were added to the HEPES buffer. The mixture was stirred for 3 h at 4 °C and further centrifuged at 2,000×g for 10 min. The 5% dextran-treated HEPES buffer was added to 500 mL of FBS and kept for 3 h at room temperature with gentle stirring. The mixture was collected and the above step was repeated overnight at 4 °C. The mixture was centrifuged at 8,000×g for 10 min at 4 °C and further filtered through nitrocellulose membrane (0.2 µm) filter (Millipore, Germany). The filtered FBS were aliquoted and stored at −20 °C until further use.
Generation of tamoxifen-resistant cells
MCF-7 cells were cultured continuously in phenol red free DMEM supplemented with 10% CSS, 1% penicillin/streptomycin and 0.1% Plasmocin (Invivogen, USA). The cells were treated with TAM (1 µM) continuously and the media were changed on alternate days. After the initial first week of treatment with TAM, 95–98% of cells underwent cell death. During 8 weeks of treatment with TAM, the cells were grown in phenol red free media and they started to develop tamoxifen resistance (TR). The treatment with TAM was continued up to 6 months and the cells were found to develop TR.
Colony formation assay
MCF-7 and MCF7-TR cells were plated in 6-well plates at a density of 500 cells/well with TAM (1 µM) and without TAM for 7 days. After 7 days, media were discarded and both MCF-7 and MCF7-TR cells were stained with crystal violet.
Cell proliferation assay
MCF-7 and MCF7-TR cells were plated in 96-well plates at a density of 1×104 cells/well. After the cells were attached, the cells were incubated with TAM (1 µM). The control cells received 0.001% ethanol as a vehicle. After 48 h, the cell viability was determined by using MTT assay.
Cell cycle analysis
The cells were treated with TAM (1 µM). The cells were washed with PBS and fixed in ice-cold 70% ethanol and kept at 4 °C overnight. Subsequently, the cells were washed with PBS twice and treated with Ribonuclease A (100 µg/mL) for 5 min at room temperature. The cells were stained with propidium iodide (PI) (50 µg/mL). The samples, the forward and side scatter, were analyzed by using a BD FACS caliber at 605 nm band pass filter. A minimum of 104 events were analyzed for each sample.
ALDEFLUOR assay
The cells were incubated with activated ALDEFLUOR reagent in ALDEFLUOR assay buffer with and without ALDH specific inhibitor diethylaminobenzaldehyde (DEAB) for 30 min in the dark at 37 °C (Stem cell technologies, Canada). The cells were further washed thrice using ALDEFLUOR assay buffer and propidium iodide (1 µg/mL) was added to assess the cell viability. ALDH inhibitor treated cells were used to establish the sorting gates, ALDH+ and ALDH− cells were sorted out using BD FACSAria flow cytometer. The ALDH+ and ALDH− cells (5,000 viable cells/mL) were cultured in ultra-low attachment plate and supplemented with MammoCultTM growth factor bullet.
Mammosphere formation assay
Mammosphere formation assay was performed as described previously (22). After ALDEFLUOR assay, the tamoxifen-sensitive and TR cells were sorted out by using fluorescence-activated cell sorting (FACS) and plated in ultra-low attachment 96-well plates at a density of 1,000 cells per well in MammoCultTM media supplemented with and without TAM (1 µM). The number of mammospheres formed after 14 days of culture was counted under light microscope.
Quantitative real-time polymerase chain reaction (QPCR)
MCF-7 and MCF7-TR cells were lysed and processed for total RNA isolation by using the Qiagen RNeasy Mini kit (Qiagen, Germany). The isolated total RNA was treated with RNase-free DNase enzyme (BioRad). The total RNA integrity was determined by using microfluidics based agarose gel electrophoresis Bioanalyzer chip (Bioanalyzer 2100, Agilent Technologies, USA). The isolated total RNA was converted to cDNA using reverse transcriptase enzyme (iScriptTM cDNA synthesis kit, BioRad, USA). The template cDNA (2 µg) was used to determine the expression profiles of the multidrug resistance genes, Cancer Drug Resistance and Metabolism PCR Array. The customized histone demethylase family regulators and stem cell marker genes were analyzed according to the protocols recommended by the manufacturer (Qiagen, Germany).
Chromatin immunoprecipitation (ChIP) assay
The MCF-7 and MCF7-TR cells were cultured in 15 cm2 petri plates until 80% of confluence was attained. The cells were trypsinized and viability was determined by using the trypan blue exclusion method. The cells were treated with 0.5 M EDTA in DMEM for 30 min. The cleavage of Notch-1 intra cellular domain (NICD) and the EDTA activity were arrested by using 1 mM glycine for 15 min. Ice-cold Phosphate Buffered Saline was used to wash the cell pellets which was collected for sonication. The chromatin was fragmented into 100–200 bp. The chromatin fragments were incubated with mouse monoclonal NICD antibody (Santa Cruz Biotechnology, USA) immunoprecipitation by using magnetic beads (Dynabeads) in accordance with the manufacturer’s protocol (Thermofisher, USA). Briefly, spin columns with magnetic beads were washed twice with wash buffer (1×). Subsequently, the reagents were added to the spin columns in the ensuing order: wash buffer (1×), cell lysate, specific primary antibody against Notch intracellular domain or negative control antibody IgG. The immunoprecipitation reactions were carried out at 4 °C in overnight. In the subsequent day, the columns were washed thrice using wash buffer and from the columns proteins were eluted by denaturing elution buffer containing β-mercaptoethanol. The eluted DNA was used for real-time PCR amplification of cyclin D.
Western blot analysis
The cells were washed twice with ice-cold PBS (pH 7.4) and then lysed with RIPA buffer (150 mmol/L NaCl, 50 M Tris, 1 mmol/L EDTA, 1% NP-40, 0.5% sodium deoxycholate, and 0.1% SDS, pH 7.4) composed of protease inhibitor cocktail (Sigma-Aldrich, USA). Cell lysates were then clarified by centrifugation at 12,000×g for 10 min at 4 °C. The total protein was quantified by using Bradford’s method (BioRad, USA). Eighty microgram (80 µg) of the total protein was subjected to SDS–PAGE (12%), and the resolved proteins were then transferred onto a PVDF membrane (BioRad, USA). The membrane was then incubated overnight at 4 °C with primary antibodies of human ERα, Notch-1, and CD44 in accordance with the manufacturer’s recommendations (1:500 to 1:1,000). After the incubation, the primary antibodies were removed and washed thrice with PBST in room temperature. The mouse/goat/anti-rabbit horseradish peroxidase-conjugated (HRP) secondary antibodies were diluted (1:10,000) in 5% of non-fat dry milk and incubated for 1 h at room temperature with gentle rocking. After washing thrice with PBST, the ECL chemical (BioRad, USA) reagents (1:1 ratio) were added to the membrane. The chemiluminescence was captured by using Bio-Rad chemiDoc imager at different time points. The equal loading/internal control β-actin was used and the band intensities of the proteins were normalized by using QuantityOne software (BioRad, USA).
Statistical analysis
All the data mean ± SEM were analyzed statistically by using one-way ANOVA followed by post hoc Student-Newman-Keuls test by using the SPSS software 7.5, which was used to compare among the groups. The statistical significance was considered at P<0.05.
Results
Generation and characterization of TR cells
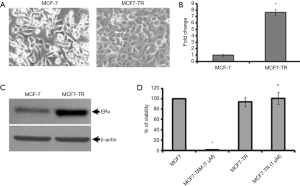
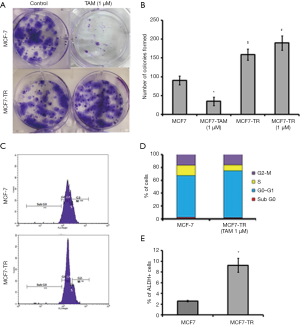
The MCF-7 cells were treated with TAM (1 µM) for six months and it resulted in the generation of TR clones, MCF7-TR cells (Figure 1A). The MCF7-TR cells were cultured continuously in phenol red free and steroid free media. Overexpression of estrogen receptor α (ERα) in MCF7-TR cells were confirmed by using QRT–PCR and western blot analyses when compared with parental MCF-7 cells (Figure 1B,C). The treatment with TAM did not alter the cell proliferation in MCF7-TR cells; however, TAM treatment reduced the cell viability of parental MCF-7 cells significantly (Figure 1D). Furthermore, the treatment with TAM increased the number of colonies in MCF7-TR cells significantly when compared with parental MCF-7 cells (Figure 2A,B). The DNA content stained by propidium iodide showed that the cell cycle progression of S phase and G2-M phase was higher in MCF7-TR cells, which indicates the cell proliferation in MCF7-TR cells (Figure 2C,D).
TAM treatment increases stem-like cells
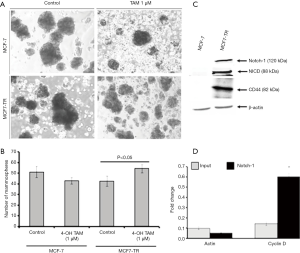
The results from the ALDEFLUOR assay showed that MCF7-TR cells have a higher number of ALDH+ cells (9.24%) than MCF-7 (2.58%) (Figure 2E), which indicate that the treatment with TAM increases stem-like phenotypes in breast cancer cells. Interestingly, the mammosphere formation was not affected in MCF-7 parental cells even when treated with TAM. However, a significant increase in the number of mammospheres in the presence of TAM (1 µM) was produced by MCF7-TR cells when compared with respective controls (Figure 3A,B). The ALDH– cells did not form mammospheres, which is in consistent with our previous study (22). This experiment suggests that the mammospheres were composed of stem-like cells and do not express estrogen receptors. This might be a reason that the stem-like cells do not respond to the treatment with TAM. In addition, the expression of Notch-1 was higher in MCF7-TR cells when compared with the parental MCF-7 cells (Figure 3C). In addition, the Notch intracellular domain (NICD) was higher in TR cells than in MCF-7 cells significantly (Figure 3C). Furthermore, CD44 expression was higher in MCF7-TR cells than in the control cells (Figure 3C). These results confirmed that the long-term treatment with TAM increased the Notch-1 mediated signaling in TR cells. Further analysis using Notch-1 antibodies with ChIP, quantitative RT-PCR showed that cyclin D was significantly higher in MCF7-TR cells due to TAM that induced the gene transcription of Notch-1 (Figure 3D). These results revealed that TR induces cell cycle progression and epigenetic changes, which increases the stem cell signaling in breast cancer cells.
TAM treatment induces the epigenetic changes associated with the development of MDR
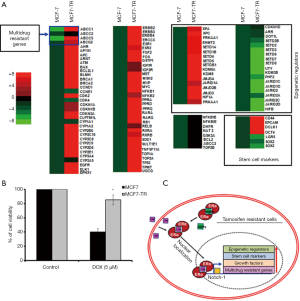
We performed 123 gene expression analysis using the MCF-7 and MCF7-TR cells. The treatment with TAM resulted in the overexpression of histone demethylase, MDR genes, cell cycle regulators and stem cell marker genes (Figure 4A). Furthermore, the expression of 23 genes was under-regulated and 7 genes did not alter (Figure 4A). The customized PCR array results suggested that several ATP-binding cassette family genes, ABCC1, ABCC2, ABCC3 and ABCG2, were upregulated significantly, confirming the development of MDR in MCF7-TR cells than in parental MCF-7 cells. In addition, we found that some stem cell marker genes, such as CD44, EPCAM, OCT4 and DCLK1, were overexpressed significantly in MCF7-TR cells than in parental MCF-7 cells, indicating the development of stem-like phenotypes in MCF7-TR cells. Furthermore, we analyzed the gene expression of 28 epigenetic histone demethylase in MCF7-TR cells, which showed that 12 histone demethylases were overexpressed (EHMT2, SETD1A, SETD1B, SETD4, SETD5, SETDB1, KDM6A, KDM8, JMJD4, JARID1A, JMJD2B, JMJD3) and the others were underexpressed (DOT1L, SETDB2, SETD2, SETD3, SETD6, SETD7, SETD8, UTY, KDM3B, PHF2, JARID1C, JARID1D, JARID2, JMJD2A, JMJD2C, JMJD2D). The heat map indicates clearly that the gene expression status was altered in MCF7-TR cells than in MCF-7 parental cells (Figure 4A).
MCF7-TR cells develop resistant to doxorubicin
MCF-7 and MCF7-TR cells were cultured with and without doxorubicin (5 µM) for 48 h. The cell survival was significantly higher in MCF7-TR cells when compared with MCF-7 cells. However, doxorubicin treatment killed MCF7-TR cells slightly but it is not significant when compared with untreated MCF-7 and MCF7-TR cells (Figure 4B). This result demonstrates that TR cells have acquired epigenetic changes associated with the overexpression of ABC transporters and stem-like genes that leads to MDR in breast cancer cells (Figure 4C).
Discussion
Our present study suggested that the treatment with TAM can alter the epigenetic regulators, which then could induce the expression of stem-like genes and the development of MDR in ERα+ breast cancer cells. We established TR in MCF-7 cells by continuing the treatment with TAM for 6 months. Our ALDEFLUOR assay data showed that TR cells have a higher number of stem-like cells. Furthermore, our study demonstrated that the MCF7-TR-derived ALDH1+ cells formed a higher number of mammospheres than MCF-7-derived mammospheres. In this context, a recent study by Wang et al. (23) suggested that ERα36 plays an important role in TR and induces stem-like phenotypes and the expression of ALDH1. However, isoform ERα66 expression was found to be decreased in estrogen receptor-activated stemness in breast cancer cells (24). Our study showed that MCF7-TR cells have higher Notch-1 expression, which could activate stem cell signaling and induce cell proliferation through cyclin D1 genes that can result in cell proliferation. Consistent with our findings, Hao et al. (25) have shown that estrogen receptor signaling activates Notch-1 signaling through IKKα in breast cancer cells. Estrogen-resistant breast cancer cells have ALDH1 activity which increases the NOTCH4 signaling that leads to a higher number of breast cancer stem cells (26). A crosstalk between Notch4 and PKCα in estrogen resistance breast cancer has also been demonstrated previously (27).
In the present study, the expression of 28 epigenetic histone demethylase genes was analyzed in MCF7-TR cells, which showed that 12 histone demethylases were overexpressed (EHMT2, SETD1A, SETD1B, SETD4, SETD5, SETDB1, KDM6A, KDM8, JMJD4, JARID1A, JMJD2B, JMJD3) and the others were underexpressed. Our results have shown a 248-fold overexpression of JMJD2B/KDM4B in ER+ MCF7-TAMR cells when treated with TAM. A recent study (28) has shown that SERM induced the expression of F-box-mediated complex degradation of JMJD2/KDM4B. Studies have shown that SETDB1 was overexpressed almost 50-fold in MCF7-TR cells and this could induce the epithelial to mesenchymal transition (EMT) and induce metastasis in breast cancer cells (29,30). JMJD3 was overexpressed (12-fold) in MCF7-TR cells and it could be involved in the regulation of stem cell genes. In contrast, a recent study (31) has suggested that the ectopic overexpression of JMJD3 may lead to reduced stem-like character and reduced OCT-4 genes. EHMT2 is a metastatic regulator overexpressed in TR cells and it could induce cell migration and metastasis. In addition, EHMT2 regulating E-cadherin, Claudin 1 and Vimentin induced EMT by activating invasion and migration of breast cancer cells (32).
KDM3B overexpression associated with better prognosis in breast cancer cells has already been reported (33). However, MCF7-TR cells have shown underexpression of KDM3B (425-fold) in TR condition, which may lead to advanced types of cancer. JMJD2D was involved in the regulation of transcription factors, such as estrogen and androgen receptors (34). The present study showed that JMJD2D expression was downregulated by 95-fold in MCF7-TR cells, and it could be TAM that modulates the expression of JMJD2D in breast cancer cells.
Our results have also shown that the gene expression of stem cell markers, such as CD44, DCLK1, OCT-4 and ABCG2 was higher in MCF7-TR cells significantly, which indicates that treatment with TAM increases the stem-like phenotypes. In addition, ALDH1 was found to be expressed highly in MCF7-TR cells when compared with parental MCF-7 cells. In consistent with the previous studies (6,23), the treatment with TAM increases the stem-like phenotypes in breast cancer cells. Interestingly, the expression of EPCAM and DCLK-1 genes was higher in MCF7-TR cells when compared with MCF-7 parental cells.
The MCF7-TR cells have higher expression of MDR genes, such as ABCC1, ABCC3, ABCC5 and ABCG2 and lead to multidrug resistance. Furthermore, chemotherapeutic drug doxorubicin did not kill MCF7-TR cells. Previous studies (4,12,35) have also supported the notion that the treatment with TAM induces the expression of ABC transporter in breast cancer cells and lead to the development of MDR in breast cancer cells. Turner et al. (36) demonstrated the FGF signaling associated with TAM resistance. Consistently, our data have also shown that the expression of FGF increased many folds in MCF7-TR cells. CYP2B6 polymorphism was observed in early hormone receptor-positive breast cancers (37). Our results showed an overexpression of CYP2B6 and it could be an increased copy number and associated with the overexpression of CYP2B6 gene in MCF7-TR cells.
Conclusions
The present study provides the evidence that treatment with TAM alters various signaling molecules epigenetically, induces stem-like phenotypes and develop MDR in breast cancer cells. This study suggests that the first-line treatment with TAM in ERα positive breast cancers could develop as MDR cancers. Therefore, targeting the epigenetic regulators may alter the development of MDR potentially and stem cell-like phenotypes in breast cancer cells.
Acknowledgments
We would like to thank Mr. M. Vijayaraman for editing the manuscript.
Funding: We gratefully acknowledge the financial assistance from the Department of Science and Technology, Science & Engineering Research Board (SERB) (Grant no. EEQ/2017/000567 dated 10 July 2018) and SRM Institute of Science and Technology internal grant (dated December 4, 2017).
Footnote
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/sci-2020-020). The authors have no conflicts of interest to declare.
Ethical Statement: The authors are accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. This study did not entail any human or animal experiments, therefore this study did not require any ethical clearance.
Open Access Statement: This is an Open Access article distributed in accordance with the Creative Commons Attribution-NonCommercial-NoDerivs 4.0 International License (CC BY-NC-ND 4.0), which permits the non-commercial replication and distribution of the article with the strict proviso that no changes or edits are made and the original work is properly cited (including links to both the formal publication through the relevant DOI and the license). See: https://creativecommons.org/licenses/by-nc-nd/4.0/.
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68:394-424. [Crossref] [PubMed]
- Jordan VC, Brodie AM. Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids 2007;72:7-25. [Crossref] [PubMed]
- Sengupta S, Jordan VC. Selective estrogen modulators as an anticancer tool: mechanisms of efficiency and resistance. Adv Exp Med Biol 2008;630:206-19. [Crossref] [PubMed]
- Sensorn I, Sukasem C, Sirachainan E, et al. ABCB1 and ABCC2 and the risk of distant metastasis in Thai breast cancer patients treated with tamoxifen. Onco Targets Ther 2016;9:2121-9. [PubMed]
- Hutcheson IR, Knowlden JM, Madden TA, et al. Oestrogen receptor-mediated modulation of the EGFR/MAPK pathway in tamoxifen-resistant MCF-7 cells. Breast Cancer Res Treat 2003;81:81-93. [Crossref] [PubMed]
- Moon YW, An HJ, Koo JS, et al. CD44/CD24 and aldehyde dehydrogenase 1 in estrogen receptor-positive early breast cancer treated with tamoxifen: CD24 positivity is a poor prognosticator. Oncotarget 2017;9:2622-30. [Crossref] [PubMed]
- Lacroix M, Leclercq G. Relevance of breast cancer cell lines as models for breast tumours: an update. Breast Cancer Res Treat 2004;83:249-89. [Crossref] [PubMed]
- Tian M, Schiemann WP. TGF-β Stimulation of EMT Programs Elicits Non-genomic ER-α Activity and Anti-estrogen Resistance in Breast Cancer Cells. J Cancer Metastasis Treat 2017;3:150-60. [Crossref] [PubMed]
- Semina SE, Scherbakov AM, Kovalev SV, et al. Horizontal Transfer of Tamoxifen Resistance in MCF-7 Cell Derivates: Proteome Study. Cancer Invest 2017;35:506-18. [Crossref] [PubMed]
- Zhang Y, Zhou G, Wang H, et al. Transcriptional upregulation of breast cancer resistance protein by 17beta-estradiol in ERalpha-positive MCF-7 breast cancer cells. Oncology 2006;71:446-55. [Crossref] [PubMed]
- Droog M, Beelen K, Linn S, et al. Tamoxifen resistance: from bench to bedside. Eur J Pharmacol 2013;717:47-57. [Crossref] [PubMed]
- Bekele RT, Venkatraman G, Liu RZ, et al. Oxidative stress contributes to the tamoxifen-induced killing of breast cancer cells: implications for tamoxifen therapy and resistance. Sci Rep 2016;6:21164. [Crossref] [PubMed]
- Vendrell JA, Magnino F, Danis E, et al. Estrogen regulation in human breast cancer cells of new downstream gene targets involved in estrogen metabolism, cell proliferation and cell transformation. J Mol Endocrinol 2004;32:397-414. [Crossref] [PubMed]
- Choi HK, Yang JW, Roh SH, et al. Induction of multidrug resistance associated protein 2 in tamoxifen-resistant breast cancer cells. Endocr Relat Cancer 2007;14:293-303. [Crossref] [PubMed]
- Cao S, Wang L, Zhang Z, et al. Sulforaphane-induced metabolomic responses with epigenetic changes in estrogen receptor positive breast cancer cells. FEBS Open Bio 2018;8:2022-34. [Crossref] [PubMed]
- Su Y, Hopfinger NR, Nguyen TD, et al. Epigenetic reprogramming of epithelial mesenchymal transition in triple negative breast cancer cells with DNA methyltransferase and histone deacetylase inhibitors. J Exp Clin Cancer Res 2018;37:314. [Crossref] [PubMed]
- Wu Q, Odwin-Dacosta S, Cao S, et al. Estrogen down regulates COMT transcription via promoter DNA methylation in human breast cancer cells. Toxicol Appl Pharmacol 2019;367:12-22. [Crossref] [PubMed]
- Cortez V, Mann M, Tekmal S, et al. Targeting the PELP1-KDM1 axis as a potential therapeutic strategy for breast cancer. Breast Cancer Res 2012;14:R108. [Crossref] [PubMed]
- Mahajan K, Lawrence HR, Lawrence NJ, et al. ACK1 tyrosine kinase interacts with histone demethylase KDM3A to regulate the mammary tumor oncogene HOXA1. J Biol Chem 2014;289:28179-91. [Crossref] [PubMed]
- Jin ML, Kim YW, Jin HL, et al. Aberrant expression of SETD1A promotes survival and migration of estrogen receptor α-positive breast cancer cells. Int J Cancer 2018;143:2871-83. [Crossref] [PubMed]
- Dandawate P, Ghosh C, Palaniyandi K, et al. The Histone Demethylase KDM3A, Increased in Human Pancreatic Tumors, Regulates Expression of DCLK1 and Promotes Tumorigenesis in Mice. Gastroenterology 2019;157:1646-59.e11. [Crossref] [PubMed]
- Palaniyandi K, Pockaj BA, Gendler SJ, et al. Human Breast Cancer Stem Cells Have Significantly Higher Rate of Clathrin-Independent and Caveolin-Independent Endocytosis than the Differentiated Breast Cancer Cells. J Cancer Sci Ther 2012;4:214-22. [Crossref] [PubMed]
- Wang Q, Jiang J, Ying G, et al. Tamoxifen enhances stemness and promotes metastasis of ERα36(+) breast cancer by upregulating ALDH1A1 in cancer cells. Cell Res 2018;28:336-58. [Crossref] [PubMed]
- Lauricella M, Carlisi D, Giuliano M, et al. The analysis of estrogen receptor-α positive breast cancer stem-like cells unveils a high expression of the serpin proteinase inhibitor PI-9: Possible regulatory mechanisms. Int J Oncol 2016;49:352-60. [Crossref] [PubMed]
- Hao L, Rizzo P, Osipo C, et al. Notch-1 activates estrogen receptor-alpha-dependent transcription via IKKalpha in breast cancer cells. Oncogene 2010;29:201-13. [Crossref] [PubMed]
- Simões BM, O'Brien CS, Eyre R, et al. Anti-estrogen Resistance in Human Breast Tumors Is Driven by JAG1-NOTCH4-Dependent Cancer Stem Cell Activity. Cell Rep 2015;12:1968-77. [Crossref] [PubMed]
- Yun J, Pannuti A, Espinoza I, et al. Crosstalk between PKCα and Notch-4 in endocrine-resistant breast cancer cells. Oncogenesis 2013;5;2:e60.
- Johmura Y, Maeda I, Suzuki N, et al. Fbxo22-mediated KDM4B degradation determines selective estrogen receptor modulator activity in breast cancer. J Clin Invest 2018;128:5603-19. [Crossref] [PubMed]
- Du D, Katsuno Y, Meyer D, et al. Smad3-mediated recruitment of the methyltransferase SETDB1/ESET controls Snail1 expression and epithelial-mesenchymal transition. EMBO Rep 2018;19:135-55. [Crossref] [PubMed]
- Yang W, Su Y, Hou C, et al. SETDB1induces epithelial mesenchymal transition in breast carcinoma by directly binding with Snail promoter. Oncol Rep 2019;41:1284-92. [PubMed]
- Xun J, Wang D, Shen L, et al. JMJD3 suppresses stem cell-like characteristics in breast cancer cells by downregulation of Oct4 independently of its demethylase activity. Oncotarget 2017;8:21918-29. [Crossref] [PubMed]
- Kim K, Son MY, Jung CR, et al. EHMT2 is a metastasis regulator in breast cancer. Biochem Biophys Res Commun 2018;496:758-62. [Crossref] [PubMed]
- Paolicchi E, Crea F, Farrar WL, et al. Histone lysine demethylases in breast cancer. Crit Rev Oncol Hematol 2013;86:97-103. [Crossref] [PubMed]
- Berry WL, Janknecht R. KDM4/JMJD2 histone demethylases: epigenetic regulators in cancer cells. Cancer Res 2013;73:2936-42. [Crossref] [PubMed]
- Baxter DE, Kim B, Hanby AM, et al. Neoadjuvant Endocrine Therapy in Breast Cancer Upregulates the Cytotoxic Drug Pump ABCG2/BCRP, and May Lead to Resistance to Subsequent Chemotherapy. Clin Breast Cancer 2018;18:481-8. [Crossref] [PubMed]
- Turner N, Pearson A, Sharpe R, et al. FGFR1 amplification drives endocrine therapy resistance and is a therapeutic target in breast cancer. Cancer Res 2010;70:2085-94. [Crossref] [PubMed]
- Kuo SH, Yang SY, You SL, et al. Polymorphisms of ESR1, UGT1A1, HCN1, MAP3K1 and CYP2B6 are associated with the prognosis of hormone receptor-positive early breast cancer. Oncotarget 2017;8:20925-38. [Crossref] [PubMed]
Cite this article as: Kalyanaraman A, Gnanasampanthapandian D, Shanmughan P, Kishore P, Ramalingam S, Arunachalam R, Jayaraman S, Kaliappan I, Munuswamy-Ramanujam G, Ramachandran I, Sambandam Y, Anbalagan M, Chandrakesan P, Palaniyandi K. Tamoxifen induces stem-like phenotypes and multidrug resistance by altering epigenetic regulators in ERα+ breast cancer cells. Stem Cell Investig 2020;7:20.


