Chromatin rules
Transcription is a key mechanism underlying the control of gene activities and cell identity during animal development and disease. Such genomic regulatory information is encoded in the form of a DNA/protein complex termed chromatin. Expression of genes underlying cell fate choices is coordinated by the binding of pioneer or lineage-specific transcription factors to gene-proximal promoters or distal enhancers. Time course measurements of transcription profiles in different cell types and their response to stimuli suggested that actively transcribed enhancers and binding of lineage-specific transcription factors coordinate for transcription events leading to cell type transition during development (1). Thus, cellular differentiation requires changes in transcription networks that are accompanied by dynamically altering local chromatin structure and enhancer/promoter activities at specific sets of genes.
Embryonic stem cells (ESCs) have the ability to differentiate into any cell type of the body. This potential is modulated by the balance of two key properties of stem cells: self-renewal and differentiation. ESCs provide an accessible in vitro model that can be biochemically and genetically manipulated to understand basic differentiation decisions such as specification of germ layers and subsequent lineage differentiation. Ability of pluripotent stem cell to differentiate into different cell types is thought to be coordinated by transcription decisions and extrinsic signaling cues. In human ESCs, genome-wide studies revealed two classes of enhancers with different chromatin signatures in early embryonic development (2). The first class of enhancers (class I) is active in stemness stage that is characterized by overlapping H3K4me1 and H3K27ac marks and required for hESC maintenance. In contrast, the second class enhancers are poised enhancers (class II) that are distinguished by the absence of H3K27ac and are required for differentiation into descendent lineages during embryonic development, but are inactive in hESCs (2). Thus, a central question is how stem cells render differentiation competence and how transcription signaling and extrinsic signals incorporate to determine cell fates.
In recent issue of Cell Stem Cell, Wang et al. described a novel enhancer epigenetic priming mechanism for acquiring developmental proficient to enable cells in response to extrinsic differentiation stimuli (3). In this study, the authors mapped the enhancer chromatin state over a time course of hESC endodermal differentiation through multiple developmental stages into terminal pancreatic and hepatic precursors. By comparing histone modifications and Gro-seq nascent transcript profiles at different stages, they revealed that H3K4me1 defines a developmental poised configuration for differentiation competent enhancers. Monomethylation of H3K4 enables these regulatory elements to respond to developmental signaling cues and to be activated in descendent lineages. Poised chromatin configuration (e.g., H3K4me1) at enhancers may be actively acquired by the binding of pioneer transcription factor Forkhead box protein A1 (FoxA1). The presence of H3K4me1 modification at lineage-specific enhancers determines cell identity, allows lineage-specific transcription factor to recognize them in response to extrinsic signaling cues, and becomes activated by subsequent H3K27 acetylation. Following enhancer activation and gene transcription, cells transit from developmental competence state into lineage-specific differentiation (Figure 1). This regulatory mechanism can occur in different stages of differentiation (Figure 1).
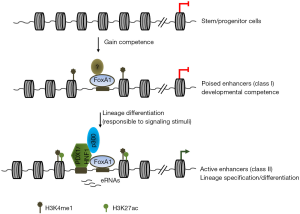
Pioneer transcription factors play a critical role in reprograming one type of cells into another (4). Such pioneer factors must have remarkable ability to recognize and engage lineage-specific genes that are developmentally silenced in stem cells (e.g., ESCs). These developmentally silenced genes are often embedded in “closed” chromatin structure that is marked by repressive histone modifications and occupied by regular nucleosomes. FoxA is a member of the forkhead class of DNA-binding transcription factors that regulate metabolic pathways and differentiation of the pancreas and liver lineages. FoxA functions as a pioneer transcriptional activator for pancreatic or liver-specific gene expression by interacting with and opening closed chromatin to initiate differentiation events. Wang et al. showed that binding of FoxA1 positively correlates with H3K4me1 and gain of developmental proficient state. Surprisingly, FOXA1 depletion did not affect the levels of H3K4me1 suggesting potential compensation effects of other forkhead transcription factors. It is unclear if the competent state of cells is affected by the FOXA1 KD (3). Testing the model that FoxA1 or other forkhead transcription factors set up H3K4me1 at poised enhancer will require additional experiments to determine whether FOXA1 directly recruits Set1/MLL histone H3K4 methyltransferase complexes or the recruitment is mediated by interaction partners of FOXA1. It is reported that FOXA1 protein physically interacts with upstream stimulatory factors (USFs) which can bind to a consensus DNA sequence similar to FOXA1 (5). USFs are also able to maintain open chromatin state by recruiting Set1/MLL and chromatin remodeling complexes to regulate ES cell pluripotency and differentiation (6,7). Thus, it is conceivable that in specific context USFs can function as pioneer transcription factors or do so by interacting with pioneer factors. Furthermore, long non-coding RNAs also could act as epigenetic regulators to control ESC pluripotency and germ layer differentiation by recruiting chromatin modifying factors to remodel local chromatin structure (8,9). Except for determining the regulatory mechanisms by which chromatin binding factors and RNA are involved in committing regulatory elements of the developmental potency programs during differentiation, equally important question is what are the regulatory mechanisms that lead to decommission of regulatory elements of stem cell programs?
Histone modifications play a crucial role in gene transcriptional regulation and in specifying cell fate decisions during development. In the stem cells, the promoters that drive expression of lineage specific genes are bookmarked by bivalent histone modifications, H3K4me3 and H3K27me3, which render developmental genes primed for differentiation program (10). However, it is well known that enhancers exert spatial and temporal controls of developmentally regulated gene expression. To determine the chromatin state of enhancers that possesses developmental proficiency for lineage differentiation, the authors examine poised enhancers in multiple developmental steps. They demonstrated that H3K4me1 primed enhancers provide a developmental competency of stem cells in multiple development steps including terminal differentiation (3). These data suggest that histone modifications play a key role in committing and coordinating enhancers and promoter to respond to developmental signals. Thus, it is interesting to test whether the poised enhancers and bivalent promoters communicate with each other to instruct specific differentiation pathways.
Finally, the authors showed that binding of lineage specific transcription factor PDX1 at the poised enhancers drives enhancer activation (e.g., H3K4me1 and H3K27ac) and pancreatic differentiation programs (3). Consistent with this observation, in the molecular level PDX1 recruits p300 to regulate pancreatic lineage specific gene expression presumably acetylating enhancer histone and leading gene activation (11). These findings provide evidence that chromatin state of DNA regulatory elements bestows cell’s ability to interpret extrinsic induction signals from their environment and determine differentiation program. In this regard, it will be particularly interesting to identify the specific chromatin-associated enzyme complexes decommissioning as well as subsequently committing the enhancers/promoters of pluripotent programs and activating them during development.
Acknowledgements
Funding: This work was supported by grants from the NIH (R56DK101994) and from the UF Health Cancer Center bridge funding.
Footnote
Conflicts of Interest: The authors have no conflicts of interest to declare.
References
- Arner E, Daub CO, Vitting-Seerup K, et al. Transcribed enhancers lead waves of coordinated transcription in transitioning mammalian cells. Science 2015;347:1010-4. [PubMed]
- Rada-Iglesias A, Bajpai R, Swigut T, et al. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011;470:279-83. [PubMed]
- Wang A, Yue F, Li Y, et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 2015;16:386-99. [PubMed]
- Iwafuchi-Doi M, Zaret KS. Pioneer transcription factors in cell reprogramming. Genes Dev 2014;28:2679-92. [PubMed]
- Sun Q, Yu X, Degraff DJ, et al. Upstream stimulatory factor 2, a novel FoxA1-interacting protein, is involved in prostate-specific gene expression. Mol Endocrinol 2009;23:2038-47. [PubMed]
- Li X, Wang S, Li Y, et al. Chromatin boundaries require functional collaboration between the hSET1 and NURF complexes. Blood 2011;118:1386-94. [PubMed]
- Deng C, Li Y, Liang S, et al. USF1 and hSET1A mediated epigenetic modifications regulate lineage differentiation and HoxB4 transcription. PLoS Genet 2013;9:e1003524. [PubMed]
- Deng C, Li Y, Zhou L, et al. HoxBlinc RNA recruits Set1/MLL complexes to activate Hox gene expression patterns and mesoderm lineage development. Cell Rep 2016;14:103-14. [PubMed]
- Guttman M, Donaghey J, Carey BW, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature 2011;477:295-300. [PubMed]
- Voigt P, Tee WW, Reinberg D. A double take on bivalent promoters. Genes Dev 2013;27:1318-38. [PubMed]
- Qiu Y, Guo M, Huang S, et al. Insulin gene transcription is mediated by interactions between the p300 coactivator and PDX-1, BETA2, and E47. Mol Cell Biol 2002;22:412-20. [PubMed]
Cite this article as: Li Y, Huang S. Chromatin rules. Stem Cell Investig 2016;3:4.


